Popular information
Popular science background:
They captured life in atomic detail (pdf)
Populärvetenskaplig information:
De fångade livet i atomdetalj (pdf)

The Nobel Prize in Chemistry 2017
Jacques Dubochet, Joachim Frank and Richard Henderson are awarded the Nobel Prize in Chemistry 2017 for their development of an effective method for generating three-dimensional images of the molecules of life. Using cryo-electron microscopy, researchers can now freeze biomolecules midmovement and portray them at atomic resolution. This technology has taken biochemistry into a new era.
They captured life in atomic detail
Over the last few years, numerous astonishing structures of life’s molecular machinery have filled the scientific literature (figure 1): Salmonella’s injection needle for attacking cells; proteins that confer resistance to chemotherapy and antibiotics; molecular complexes that govern circadian rhythms; light-capturing reaction complexes for photosynthesis and a pressure sensor of the type that allows us to hear. These are just a few examples of the hundreds of biomolecules that have now been imaged using cryo-electron microscopy (cryo-EM).
When researchers began to suspect that the Zika virus was causing the epidemic of brain-damaged newborns in Brazil, they turned to cryo-EM to visualise the virus. Over a few months, threedimensional (3D) images of the virus at atomic resolution were generated and researchers could start searching for potential targets for pharmaceuticals.
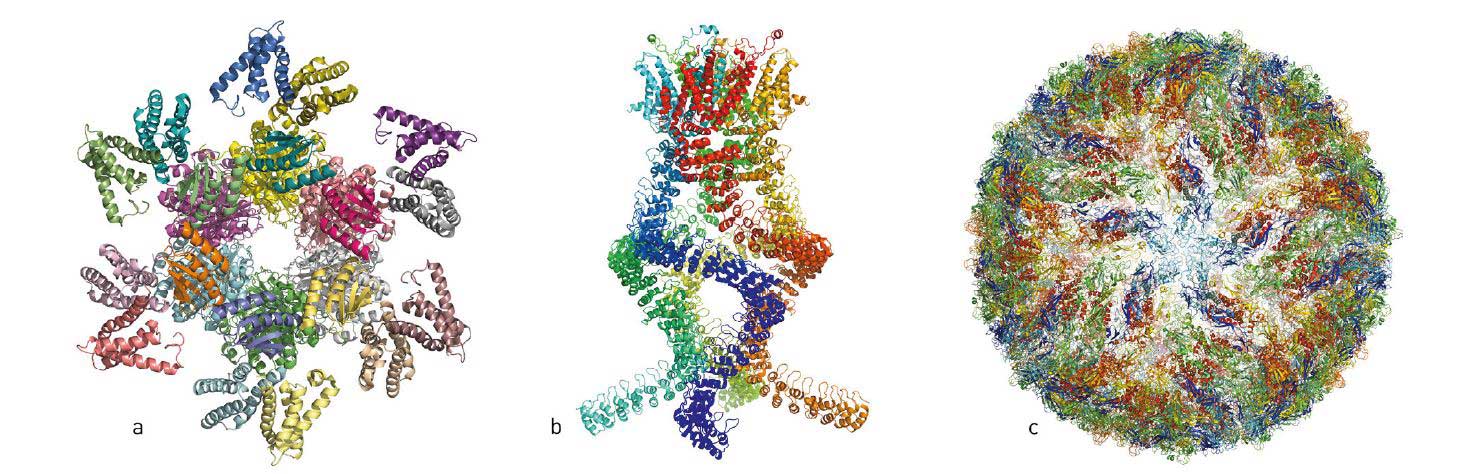
Figure 1. Over the last few years, researchers have published atomic structures of numerous complicated protein complexes. a. A protein complex that governs the circadian rhythm. b. A sensor of the type that reads pressure changes in the ear and allows us to hear. c. The Zika virus.
© Johan Jarnestad/The Royal Swedish Academy of Sciences
Jacques Dubochet, Joachim Frank and Richard Henderson have made ground-breaking discoveries that have enabled the development of cryo-EM. The method has taken biochemistry into a new era, making it easier than ever before to capture images of biomolecules.
Pictures – an important key to knowledge
In the first half of the twentieth century, biomolecules – proteins, DNA and RNA – were terra incognita on the map of biochemistry. Scientists knew they played fundamental roles in the cell, but had no idea what they looked like. It was only in the 1950s, when researchers at Cambridge began to expose protein crystals to X-ray beams, that it was first possible to visualise their wavy and spiralling structures.
In the early 1980s, the use of X-ray crystallography was supplemented with the use of nuclear magnetic resonance (NMR) spectroscopy for studying proteins in solid state and in solution. This technique not only reveals their structure, but also how they move and interact with other molecules.
Thanks to these two methods, there are now databases containing thousands of models of biomolecules that are used in everything from basic research to pharmaceutical development. However, both methods suffer from fundamental limitations. NMR in solution only works for relatively small proteins. X-ray crystallography requires that the molecules form well-organised crystals, such as when water freezes to ice. The images are like black and white portraits from early cameras – their rigid pose reveals very little about the protein’s dynamics.
Also, many molecules fail to arrange themselves in crystals, which caused Richard Henderson to abandon X-ray crystallography in the 1970s – and this is where the story of 2017’s Nobel Prize in Chemistry begins.
Problems with crystals made Henderson change track
Richard Henderson received his PhD from the bastion of X-ray crystallography, Cambridge, UK. He used the method for imaging proteins, but setbacks arose when he attempted to crystallise a protein that was naturally embedded in the membrane surrounding the cell.
Membrane proteins are difficult to manage. When they are removed from their natural environment – the membrane – they often clump up into a useless mass. The first membrane protein that Richard Henderson worked with was difficult to produce in adequate amounts; the second failed to crystallise. After years of disappointment, he turned to the only available alternative: the electron microscope.
It is open to discussion whether electron microscopy really was an option at this time. Transmission electron microscopy, as the technique is called, works more or less like ordinary microscopy, but a beam of electrons is sent through the sample instead of light. The electrons’ wavelength is much shorter than that of light, so the electron microscope can make very small structures visible – even the position of individual atoms.
In theory, the resolution of the electron microscope was thus more than adequate for Henderson to obtain the atomic structure of a membrane protein, but in practice the project was almost impossible. When the electron microscope was invented in the 1930s, scientists thought that it was only suitable for studying dead matter. The intense electron beam necessary for obtaining high resolution images incinerates biological material and, if the beam is weakened, the image loses its contrast and becomes fuzzy.
In addition, electron microscopy requires a vacuum, a condition in which biomolecules deteriorate because the surrounding water evaporates. When biomolecules dry out, they collapse and lose their natural structure, making the images useless.
Almost everything indicated that Richard Henderson would fail, but the project was saved by the special protein that he had chosen to study: bacteriorhodopsin.
The best so far was not good enough for Henderson
Bacteriorhodopsin is a purple-coloured protein that is embedded in the membrane of a photosynthesising organism, where it captures the energy from the sun’s rays. Instead of removing the sensitive protein from the membrane, as Richard Henderson had previously tried to do, he and his colleague took the complete purple membrane and put it under the electron microscope. When the protein remained surrounded by the membrane it retained its structure; they covered the sample’s surface with a glucose solution that protected it from drying out in the vacuum.
The harsh electron beam was a major problem, but the researchers made use of how the bacteriorhodopsin molecules are packed in the organism’s membrane. Instead of blasting it with a full dose of electrons, they had a weaker beam flow through the sample. The image’s contrast was poor and they could not see the individual molecules, but they used the fact that the proteins were regularly packed and oriented in the same direction. When all the proteins diffract the electron beams in an almost identical manner, they were able to calculate a more detailed image based on the diffraction pattern – they used a similar mathematical approach to that used in X-ray crystallography.
At the next stage, the researchers turned the membrane under the electron microscope, taking pictures from many different angles. This way, in 1975 it was possible to produce a rough 3D model of bacteriorhodopsin’s structure (figure 2), which showed how the protein chain wiggled through the membrane seven times.
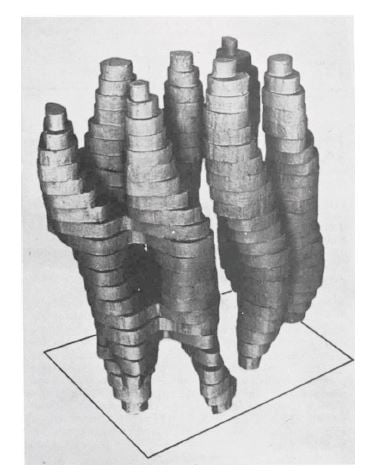
Figure 2. The first rough model of bacteriorhodopsin, published in 1975.
Image from Nature 257: 28-32
It was the best picture of a protein ever generated using an electron microscope. Many people were impressed by the resolution, which was 7 Ångström (0.0000007 millimetres), but this was not enough for Richard Henderson. His goal was to achieve the same resolution as that provided by X-ray crystallography, about 3 Ångström, and he was convinced that electron microscopy had more to give.
Henderson produces the first image at atomic resolution
Over the following years, electron microscopy gradually improved. The lenses got better and cryotechnology developed (we will return to this), in which the samples were cooled with liquid nitrogen during the measurements, protecting them from being damaged by the electron beam.
Richard Henderson gradually added more details to the model of bacteriorhodopsin. To get the sharpest images he travelled to the best electron microscopes in the world. They all had their weaknesses, but complemented each other. Finally, in 1990, 15 years after he had published the first model, Henderson achieved his goal and was able to present a structure of bacteriorhodopsin at atomic resolution (figure 3).

Figure 3. In 1990, Henderson presented a bacteriorhodopsin structure at atomic resolution.
© Johan Jarnestad/The Royal Swedish Academy of Sciences
He thereby proved that cryo-EM could provide images as detailed as those generated using X-ray crystallography, which was a crucial milestone. However, this progress was built upon an exception: how the protein naturally packed itself regularly in the membrane. Few other proteins spontaneously order themselves in this way. The question was whether the method could be generalised: would
it be possible to use an electron microscope to generate high-resolution 3D images of proteins that were randomly scattered in the sample and oriented in different directions? Richard Henderson believed it would be, while others thought this was a utopia.
On the other side of the Atlantic, at the New York State Department of Health, Joachim Frank had long worked to find a solution to just that problem. In 1975, he presented a theoretical strategy where the apparently minimal information found in the electron microscope’s two-dimensional images could be merged to generate a high-resolution, three-dimensional whole. It took him over a decade to realise this idea.
Frank refines image analysis
Joachim Frank’s strategy (figure 4) built upon having a computer discriminate between the traces of randomly positioned proteins and their background in a fuzzy electron microscope image. He developed a mathematical method that allowed the computer to identify different recurring patterns in the image. The computer then sorted similar patterns into the same group and merged the information in these images to generate an averaged, sharper image. In this way he obtained a number of high-resolution, two-dimensional images that showed the same protein but from different angles. The algorithms for the software were complete in 1981.
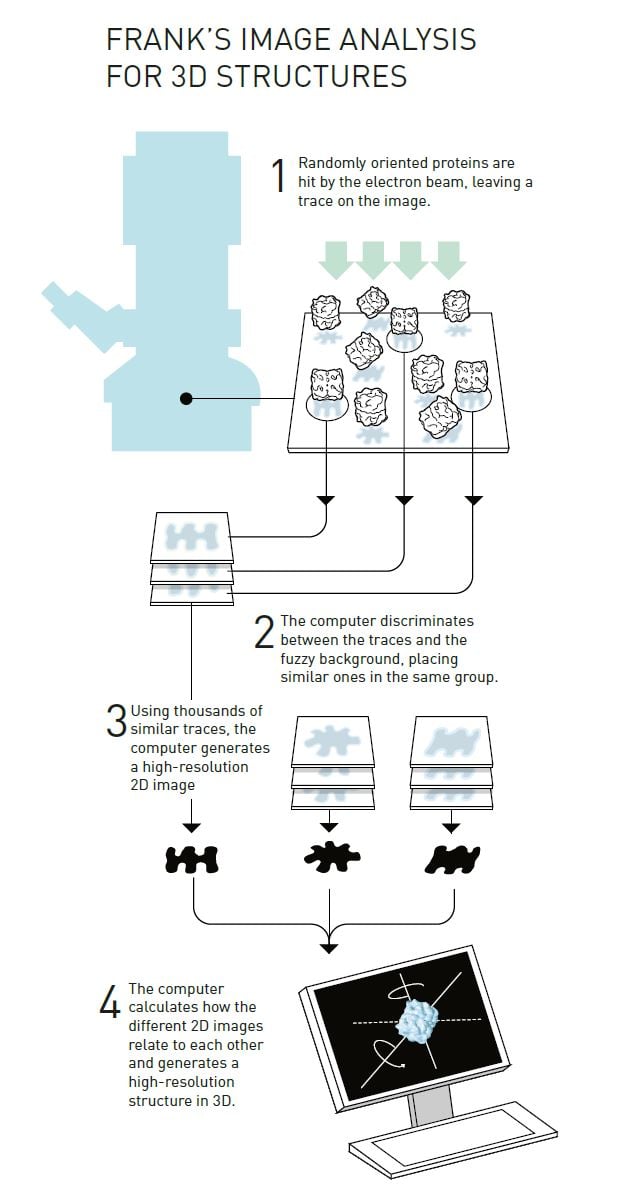
© Johan Jarnestad/The Royal Swedish Academy of Sciences
The next step was to mathematically determine how the different two-dimensional images were related to each other and, based on this, to create a 3D image. Frank published this part of the image analysis method in the mid-1980s and used it to generate a model of the surface of a ribosome, the gigantic molecular machinery that builds proteins inside the cell.
Joachim Frank’s image processing method was fundamental to the development of cryo-EM. Now we’ll jump a few years back in time – in 1978, at the same time as Frank was perfecting his computer programs, Jacques Dubochet was recruited to the European Molecular Biology Laboratory in Heidelberg to solve another of the electron microscope’s basic problems: how biological samples dry out and are damaged when exposed to a vacuum.
Dubochet makes glass from water
In 1975, Henderson used a glucose solution to protect his membrane from dehydrating, but this method did not work for water-soluble biomolecules. Other researchers had tried freezing the samples because ice evaporates more slowly than water, but the ice crystals disrupted the electron beams so much that the images were useless.
The vaporising water was a major dilemma. However, Jacques Dubochet saw a potential solution: cooling the water so rapidly that it solidified in its liquid form to form a glass instead of crystals. A glass appears to be a solid material, but is actually a fluid because it has disordered molecules.
Dubochet realised that if he could get water to form glass – also known as vitrified water – the electron beam would diffract evenly and provide a uniform background.
Initially, the research group attempted to vitrify tiny drops of water in liquid nitrogen at –196°C, but were successful only when they replaced the nitrogen with ethane that had, in turn, been cooled by liquid nitrogen. Under the microscope they saw a drop that was like nothing they had seen before. They first assumed it was ethane, but when the drop warmed slightly the molecules suddenly rearranged themselves and formed the familiar structure of an ice crystal. It was a triumph – particularly as some researchers had claimed it was impossible to vitrify water drops. We now believe that vitrified water is the most common form of water in the universe.
A simple technique for contrast
After the breakthrough in 1982, Dubochet’s research group rapidly developed the basis of the technique that is still used in cryo-EM (figure 5). They dissolved their biological samples – initially different forms of viruses – in water. The solution was then spread across a fine metal mesh as a thin film. Using a bow-like construction they shot the net into the liquid ethane so that the thin film of water vitrified.
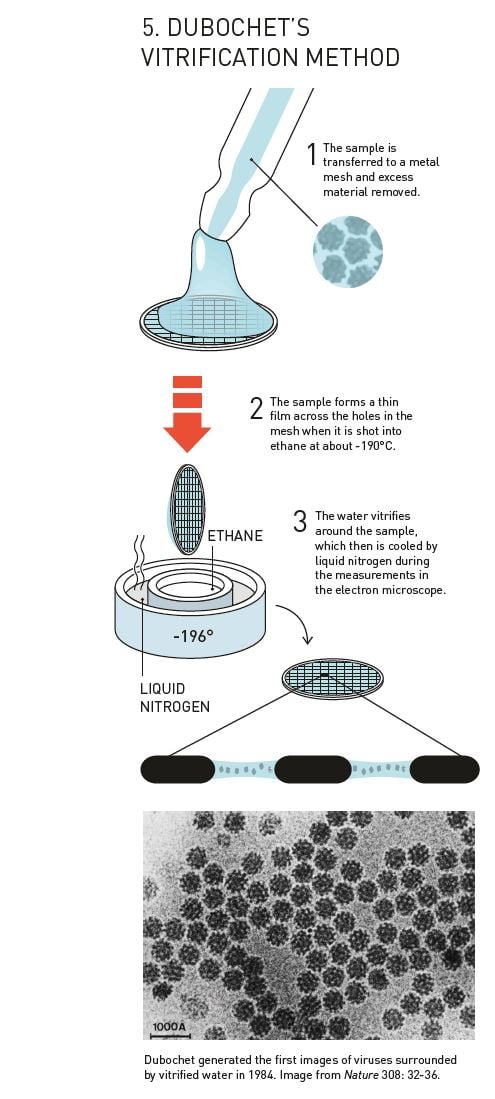
In 1984, Jacques Dubochet published the first images of a number of different viruses, round and hexagonal, that are shown in sharp contrast against the background of vitrified water. Biological material could now be relatively easily prepared for electron microscopy, and researchers were soon knocking on Dubochet’s door to learn the new technique.
From blobology to revolution
The most important pieces of cryo-EM were thus in place, but the images still had poor resolution. In 1991, when Joachim Frank prepared ribosomes using Dubochet’s vitrification method and analysed
the images with his own software, he obtained a 3D structure that had a resolution of 40 Å. It was an amazing step forward for electron microscopy, but the image only showed the ribosome’s contours. Frankly, it looked like a blob and the image did not even come close to the atomic resolution of X-ray crystallography.
Because cryo-EM could rarely visualise anything other than an uneven surface, the method was sometimes called “blobology”. However, every nut and bolt of the electron microscope has gradually been optimised, greatly due to Richard Henderson stubbornly maintaining his vision that electron microscopy would one day routinely provide images that show individual atoms. Resolution has improved, Ångström by Ångström, and the final technical hurdle was overcome in 2013, when a new type of electron detector came into use (figure 6).
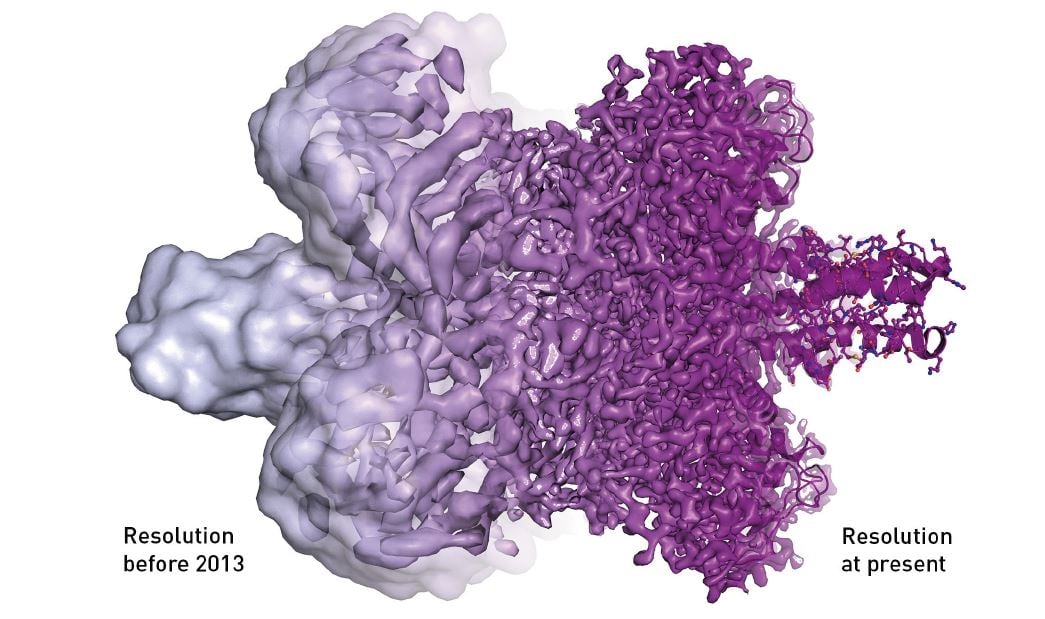
Figure 6. The electron microscope’s resolution has radically improved in the last few years, from mostly showing shapeless blobs to now being able to visualise proteins at atomic resolution.
© Martin Högbom/The Royal Swedish Academy of Sciences
Every hidden corner of a cell can be explored
Now the dream is reality, and we are facing an explosive development within biochemistry. There are a number of benefits that make cryo-EM so revolutionary: Dubochet’s vitrification method is relatively easy to use and requires a minimal sample size. Due to the rapid cooling process, biomolecules can be frozen mid-action and researchers can take image series that capture different parts of a process. This way, they produce ‘films’ that reveal how proteins move and interact with other molecules.
Using cryo-EM, it is also easier than ever before to depict membrane proteins, which often function as targets for pharmaceuticals, and large molecular complexes. However, small proteins cannot be studied with electron microscopy, but they can be visualised using NMR spectroscopy or X-ray crystallography.
After Joachim Frank presented the strategy for his general image processing method in 1975, a researcher wrote: “If such methods were to be perfected, then, in the words of one scientist, the sky would be the limit.”
Now we are there – the sky is the limit. Jacques Dubochet, Joachim Frank and Richard Henderson have, through their research, brought “the greatest benefit to mankind.” Each corner of the cell can be captured in atomic detail and biochemistry is all set for an exciting future.
LINKS AND FURTHER READING
Additional information on this year’s prizes, including a scientific background in English, is available on the website of the Royal Swedish Academy of Sciences, www.kva.se, and at http://nobelprize.org. There you can watch video footage of the press conferences, the Nobel Lectures and more. Information on exhibitions and activities related to the Nobel Prizes and the Prize in Economic Sciences is available at www.nobelmuseum.se.
Articles
Dubochet, J. (2016) A Reminiscence about Early Times of Vitreous Water in Electron Cryomicroscopy.
Biophys J., 110, 756-757.
Elmes, J. (2016) Interview with Richard Henderson. Times Higher Education, www.timeshighereducation.com/people/interview-richard-henderson-university-of-cambridge
Gelfand, A. (2016) The Rise of Cryo–Electron Microscopy, Biomedical Computation Review, http://biomedicalcomputationreview.org/sites/default/files/riseofc-e_bcr-spring-2016-web.pdf
Mossman, K. (2007) Profile of Joachim Frank, Proc. Natl Acad. Sci. USA, 104, 19668-19670. Wilson, R. och Gristwood, A. (2015) Jacques Dubochet.
www.embl.de/aboutus/alumni/news/news_2015/20150709_dubochet/
Videos
Serious Science (20 June 2017) Electron Cryomicroscopy — Richard Henderson
www.youtube.com/watch?v=L6U–sYUF9s
Serious Science (22 Aug. 2017) Single-Particle Electron Microscopy – Richard Henderson
www.youtube.com/watch?v=j_sfs6uwWlc
The Franklin Institute (5 July 2016) Joachim Frank — 2014 Laureate of the Franklin Institute in Life Science
www.youtube.com/watch?v=mtvht8Uyh2s
The Royal Swedish Academy of Sciences has decided to award the Nobel Prize in Chemistry 2017 to
JACQUES DUBOCHET
Born 1942 in Aigle, Switzerland. Ph.D. 1973, University of Geneva and University of Basel, Switzerland.
Honorary Professor of Biophysics, University of Lausanne, Switzerland.
www.unil.ch/dee/en/home/menuinst/ people/honorary-professors/prof- jacques-dubochet.html
JOACHIM FRANK
Born 1940 in Siegen, Germany. Ph.D. 1970, Technical University of Munich, Germany. Professor of Biochemistry and Molecular Biophysics and of Bio- logical Sciences, Columbia University, New York, USA.
http://franklab.cpmc.columbia.edu/ franklab/
and
RICHARD HENDERSON
Born 1945 in Edinburgh, Scotland. Ph.D. 1969, Cambridge University, UK. Programme Leader, MRC Laboratory of Molecular Biology, Cambridge, UK.
www2.mrc-lmb.cam.ac.uk/groups/rh15/
“for developing cryo-electron microscopy for the high-resolution structure determination of biomolecules in solution”
Science Editors: Peter Brzezinski, Gunnar von Heijne and Sara Snogerup Linse, the Nobel Committee for Chemistry
Text: Ann Fernholm
Translation: Clare Barnes
Illustrations: © Johan Jarnestad/The Royal Swedish Academy of Sciences (unless otherwise credited)
Editor: Ann Fernholm
© The Royal Swedish Academy of Sciences
Nobel Prizes and laureates
Six prizes were awarded for achievements that have conferred the greatest benefit to humankind. The 14 laureates' work and discoveries range from quantum tunnelling to promoting democratic rights.
See them all presented here.
