Popular information
Information for the public:
How the jellyfish’s green light revolutionised bioscience (pdf)
Populärvetenskaplig information:
Manetens gröna ljus revolutionerade biovetenskapen (pdf)

The Nobel Prize in Chemistry 2008
In the 1960s, when the Japanese scientist Osamu Shimomura began to study the bioluminescent jellyfish Aequorea victoria, he had no idea what a scientific revolution it would lead to. Thirty years later, Martin Chalfie used the jellyfish’s green fluorescent protein to help him study life’s smallest building block, the cell. Today, scientists are able to study biological processes that were previously invisible with the aid of Roger Y. Tsien’s proteins, which glow in all colours of the rainbow.
How the jellyfish’s green light revolutionised bioscience
When scientists develop methods to help them see things that were once invisible, research always takes a great leap forward. For example, when Anton van Leeuwenhoek invented the microscope in the 17th century a new world opened up. Scientists could suddenly see bacteria, sperm and blood cells. Things they previously did not know even existed.
This year’s Nobel Prize in Chemistry rewards a similar effect on science. The green fluorescent protein, GFP, has functioned in the past decade as a guiding star for biochemists, biologists, medical scientists and other researchers. The strong green colour of this protein appears under blue and ultraviolet light. It can, for example, illuminate growing cancer tumours; show the development of Alzheimer’s disease in the brain or the growth of pathogenic bacteria.
An even more interesting use of GFP means that researchers can actually follow processes inside individual cells. The body consists of billions of cells, from pumping heart muscle cells and insulin-producing beta cells to macrophages that destroy unwelcome bacteria. The more researchers know about a cell type – how it develops and functions – the greater the chance that they can develop effective drugs with minimal side-effects.
Studying the machinery of these 0.02 millimetre sized cells is not easy. Observing the building blocks of a cell: proteins, fatty acids, carbohydrates and other molecules is beyond the power of an ordinary microscope. And it is even more difficult to follow chemical processes within a cell, but it is at this detailed level that scientists must work. When researchers understand how cells start building new blood vessels, for example, they might be able to stop cancer tumours from acquiring a nourishing and oxygenating vessel system. This will prevent their growth.
The chemical processes of cells are usually regulated by proteins. There are tens of thousands of different proteins, each with different functions. By connecting GFP to one of these proteins, researchers can obtain vital information. They can see which cells a particular protein inhabits, they can follow its movements and watch its interactions with other proteins. Thanks to GFP’s green light scientists can now track a single protein under the microscope.
Shimomura fishes for luminescent material
Today, GFP is a standard tool for thousands of researchers all over the world. The story of its discovery starts in Japan in the years after the Second World War. Osamu Shimomura’s education was disrupted by the war and the devastation caused by the atom bomb. Despite this, in 1955 he was employed as an assistant by Professor Yashimasa Hirata at Nagoya University. Professor Hirata put him to work on a seemingly impossible project – to discover what made the remains of a crushed mollusc, Cypridina, glow when it was moistened with water.
It may seem strange that Hirata gave such an inexperienced assistant such a difficult task. A leading American research group had tried for a long time to isolate this material, so Hirata decided that he did not want to give the job to a student who needed to succeed in order to get his or her doctorate.
In 1956, against all odds, Shimomura had the material in his hand. It was a protein that glowed 37,000 times more brightly than the crushed mollusc. After publishing his results, Shimomura was recruited to the prestigious University of Princeton in New Jersey, USA, by Frank Johnson. As a farewell present, Professor Hirata saw to it that Shimomura was awarded a Ph.D. from Nagoya University – an unusual act, as he was not actually enrolled as a doctoral student.
After a long journey across the Pacific Ocean and the American continent, Shimomura set about studying another naturally luminescent material. This time it was from the jellyfish Aequorea victoria, whose outer edge glows green when the jellyfish is agitated.
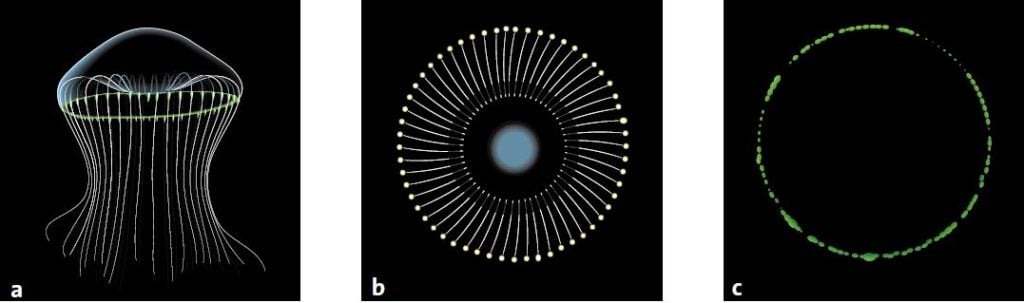
During the whole of the summer of 1961, Shimomura and Johnson gathered jellyfish in Friday Harbor on the west coast of North America. They cut off the edges of the jellyfish and pressed them through a filter to get what they called a ‘squeezate’. One day when Shimomura poured some of the squeezate into the sink, it flashed brightly. He realised that there was seawater in the sink and that it was calcium ions in the seawater that had caused the chemical reaction. Strangely enough, the flash of light was not green like the edges of the jellyfish. It was blue.
Johnson and Shimomura gathered raw material throughout that summer and took back squeezate from about 10,000 jellyfish to Princeton. It took them a few months to purify just a few milligrams of the blue luminescent material from the liquid. This protein they named aequorin.
It fluoresces green in UV light
In the 1962 scientific publication, in which Shimomura and Johnson described the process by which aequorin was obtained, they also mentioned that they had isolated a protein that was slightly greenish in sunlight, yellowish in the light from a light bulb and fluorescent green in UV light. This was the first time that anyone had described GFP. Shimomura and Johnson called it the green protein but later it was named the green fluorescent protein.
In the 1970s, Shimomura looked more closely at GFP’s fluorescence. He showed that GFP contains a special chromophore, a chemical group that absorbs and emits light. When UV light or blue light hits the GFP chromophore, it sucks up the energy in the light, it is excited. In the next phase, the chromophore gets rid of the energy. It emits light, which is now in the green wavelength.

In jellyfish, the GFP’s chromophore simply transforms the blue light from aequorin into green light. This is why the jellyfish and aequorin glow in different colours.
What is revolutionary about GFP is that the protein does not need any additives to glow, in contrast to aequorin and other bioluminescent proteins, which require a continous supply of energy rich molecules. It is enough to radiate GFP with UV light or blue light. The light enters the cells and meets GFP, which glows green. If the researchers had needed to add a chemical, they would have had to inject it into the cell – a process which can both disturb the cell and is difficult to carry out at such microscopic scales.
Chalfie has a brilliant idea
This year’s second Nobel laureate in chemistry, Martin Chalfie, heard about the green fluorescent protein for the first time in 1988 at a seminar dealing with bioluminescent organisms at Columbia University in New York. When Chalfie heard that there was a glowing protein, he was delighted.
In his everyday work, Chalfie dealt with the millimetre-long roundworm Caenorhabditis elegans, one of the most frequently studied organisms in the world. Although it consists of only 959 cells, it has a brain, it grows old and it mates. In addition, a third of the roundworm’s genes are related to human genes. Last but not least, C. elegans is transparent, which makes it easy for researchers to study its organs under an ordinary microscope.
During the 1988 seminar, Chalfie realised that green fluorescent protein would be a fantastic tool for mapping the roundworm. It would act as a glowing green signal for various activities in the roundworm’s cells.
If we are to fully appreciate Chalfie’s idea, we need to know some basic facts about cell biology. As mentioned before, various proteins carry out almost all the work in a cell and there are tens of thousands of proteins in our bodies. Although they serve so many different functions, all proteins are constructed in the same way. They consist of 20 different types of amino acids that are linked together in long chains. What distinguishes one protein from another is the length of the chain, the sequence of amino acids and how the chain folds.
In general, each gene is a description of a protein. When a protein is needed in a cell, the gene is activated, which results in the cell producing the required protein.
For instance, when you have eaten a big bag of sweets and your blood-sugar level is too high, the insulin gene in the pancreatic beta cells is switched on. All cells in the body have the insulin gene safely inside the cell nucleus, but only beta cells react to the blood-sugar level and start to produce insulin. The switch for the gene, the promoter, which lies close to the gene in the DNA, is turned on. When the promoter is active, the insulin gene begins to be copied. It is like copying a valuable old book that is kept in a fireproof storeroom. This copy is necessary if the cell needs to access and read the genetic blueprint.
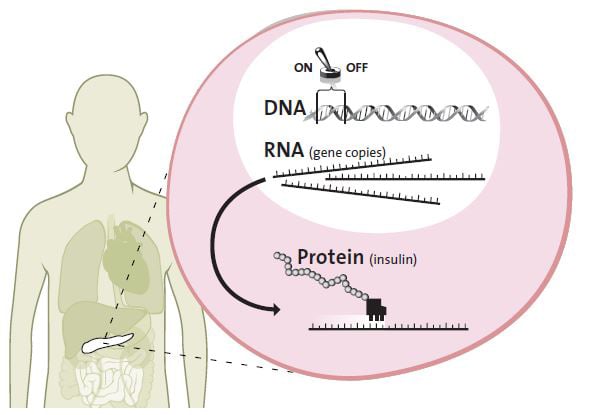
The copy of the insulin gene is transferred from the cell nucleus to the cell workshop, the cytoplasm. Then the gene copy is used as a pattern for bringing the amino acids together, forming the protein insulin. The insulin is released into the bloodstream where it sticks to muscle and fat cells, which absorb and store sugar from the blood.
Chalfie’s idea was that by connecting the gene for GFP with various gene switches or with genes from other proteins, he would be able to watch cells gene switches activate and he would be able to see where different proteins were produced. The green light would act as a beacon for various events.
An unexpected discovery
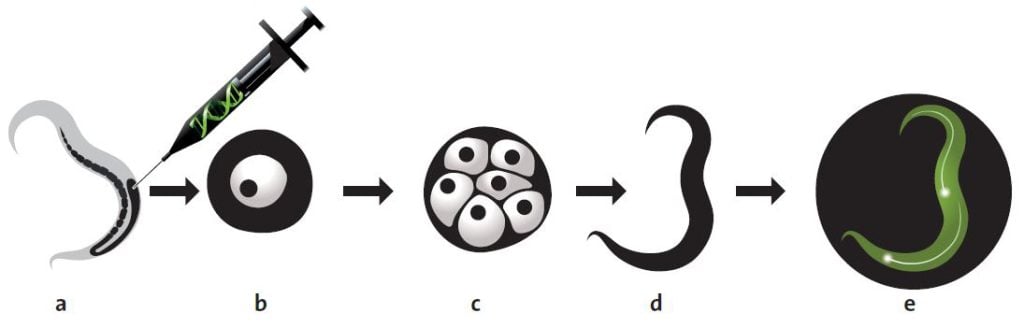
In order to test his ideas, Chalfie needed to locate the gene for GFP in the genome of Aequorea victoria. After a few investigations, Chalfie found out that a researcher called Douglas Prasher at Woods Hole Oceanographic Institution in Massachusetts had already started to look for the GFP gene. Chalfie contacted Prasher and asked him to get in touch if he succeeded in isolating the right gene. In research language this is called cloning a gene. A researcher isolates the gene from an organism’s genome and, with the help of DNA technology, places it in a single-celled organism, which is easier to manage. Usually researchers use the ordinary intestinal bacterium, Escherichia coli. They can then transform the bacterium into a protein factory by activating the foreign gene.
A couple of years later, Prasher sent the GFP gene to Chalfie. Chalfie then instructed a graduate student, Ghia Euskirchen, in ways to attempt to get E. coli to produce GFP.
About one month later, Euskirchen called Chalfie. She had succeeded! They could see in the microscope that the bacteria glowed green when they were radiated with UV light. This discovery forms the basis of today’s revolutionary use of GFP. But the discovery itself was quite unexpected.
In the early 1990’s, scientists generally assumed that naturally fluorescing molecules and pigments (which give flowers, fish and other organisms their colour) were produced in several steps in the cells. Each of these steps requires a protein to control the chemical production. Many experts believed that a few different proteins were needed to produce the chromophore in GFP, but Chalfie and Euskirchen’s experiment showed that this premise was wrong. No other protein than GFP was needed.
In the next step, Chalfie placed the gene behind a promoter that is active in six touch receptor neurons in C. elegans. The results were published by Chalfie and his colleagues in the scientific journal Science in February 1994. On the front cover, readers could see an image of C. elegans in which the touch receptor neurons were glowing bright green.
Tsien creates a palette with all the colours of the rainbow
This is where the third Nobel Prize laureate Roger Tsien makes his entry. His greatest contribution to the GFP revolution was that he extended the researchers’ palette with many new colours that glowed longer and with higher intensity.
To begin with, Tsien charted how the GFP chromophore is formed chemically in the 238-amino acid-long GFP protein. Researchers had previously shown that three amino acids in position 65–67 react chemically with each other to form the chromosphore. Tsien showed that this chemical reaction requires oxygen and explained how it can happen without the help of other proteins.
With the aid of DNA technology, Tsien took the next step and exchanged various amino acids in different parts of GFP. This led to the protein both absorbing and emitting light in other parts of the spectrum. By experimenting with the amino acid composition, Tsien was able to develop new variants of GFP that shine more strongly and in quite different colours such as cyan, blue and yellow. That is how researchers today can mark different proteins in different colours to see their interactions.
One colour, however, that Tsien could not produce with GFP was red. Red light penetrates biological tissue more easily and is therefore especially useful for researchers who want to study cells and organs inside the body.
At this point, Mikhail Matz and Sergei Lukyanov, two Russian researchers, became involved in the GFP revolution. They looked for GFP-like proteins in fluorescent corals and found six more proteins, one red, one blue and the rest green.
The desired red protein, DsRED, was unfortunately larger and heavier than GFP. DsRED consisted of four amino acid chains instead of one and was of less use as a fluorescent tag in biological processes. Tsien’s research group solved this problem, redesigning DsRED so that the protein is now stable and fluoresces as a single amino acid chain, which can easily be connected to other proteins.
From this smaller protein, Tsien’s research group also developed proteins with mouth-watering names like mPlum, mCherry, mStrawberry, mOrange and mCitrine, according to the colour they glowed. Several other researchers and companies have also contributed new colours to this growing palette. So today, 46 years after Shimomura first wrote about the green fluorescent protein, there is a kaleidoscope of GFP-like proteins which shine with all the colours of the rainbow.
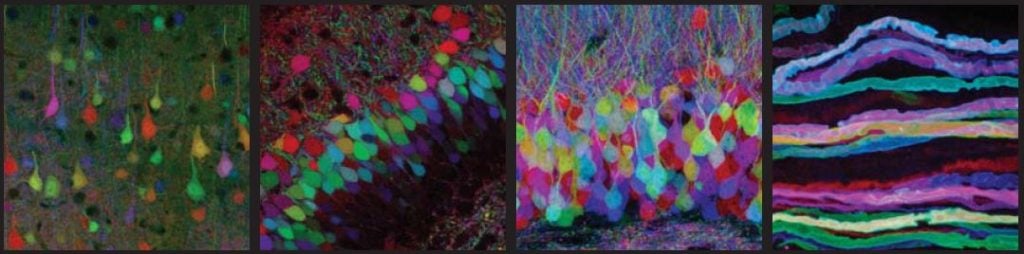
The brainbow
Three of these proteins have been used by researchers in a spectacular experiment. Mice were genetically modified to produce varying amounts of the colours yellow, cyan and red within the nerve cells of their brain. This combination of colours is similar to the one used by computer printers. The result was a mouse brain that glowed in the colours of the rainbow. The researchers could follow nerve fibres from individual cells in the dense network in the brain. The researchers called this experiment “the brainbow”.
GFP sensors for arsenic and heavy metals
The green fluorescent protein can also be used for biotechnical applications, including the detection of arsenic in water wells. This is an enormous problem in parts of South-East Asia, where naturally occurring arsenic is poisoning many thousands of people. Researchers have genetically modified arsenic-resistant bacteria that will glow green in the presence of arsenic. Scientists have also modified other organisms to fluoresce green in the presence of the explosive trinitrotoluene (TNT) or heavy metals such as cadmium or zinc. Nowadays there is even GFP in toys that glow in the dark.
One mystery remains to be solved
When Osamu Shimomura began to study biofluorescent organisms in the sea, he wanted to understand what made them shine. This is a typical example of how basic research can lead to an unexpected scientific revolution.
However, one mystery remains to be solved. Why does the jellyfish Aequorea victoria shine? Many organisms living in the sea use light from biofluorescent proteins to confuse their enemies, to attract food or to tempt a partner. But no one knows what has caused Aequoreavictoria to evolve aequorin and GFP.
LINKS AND FURTHER READING
Books
Pieribone, V., Gruber D. F., A Glow in the Dark. 2005, Cambridge, Massachusetts, and London, England, The Belknap Press, Harvard University Press
Zimmer M., Glowing Genes. 2005, Amherst, New York, Prometheus Books
Scientific review articles
Shimomura, O. (2005) The discovery of aequorin and green fluorescent protein. Journal of Microscopy 217 3-15 Shaner, N.C. et al. (2008) Improving the photostability of bright monomeric orange and red fluorescent proteins. Nature Methods 5 545-551
Link
Movie showing a cell producing GFP-tagged HIV particles (green dots):
www.nature.com/nature/journal/v454/n7201/suppinfo/nature06998.html
Website describing the GFP-revolution: www.conncoll.edu/ccacad/zimmer/GFP-ww/GFP-1.htm
THE LAUREATES
OSAMU SHIMOMURA
Marine Biological Laboratory (MBL)
7 MBL Street, Woods Hole, MA 02543, USA
and
Boston University Medical School
School of Medicine, 715 Albany Street, Boston, MA 02118, USA
www.conncoll.edu/ccacad/zimmer/ GFP-ww/shimomura.html
Japanese citizen. Born 1928 in Kyoto, Japan. Ph.D. in organic chemistry 1960, from Nagoya University, Japan.
Professor Emeritus at Marine Biological Laboratory (MBL), Woods Hole, MA, USA and Boston University Medical School, MA, USA.
MARTIN CHALFIE
Columbia University Biological Sciences
1012 Fairchild Center, M.C. 2446, New York, NY 10027 USA
www.columbia.edu/cu/biology/ faculty-data/martin-chalfie/ faculty.html
US citizen. Born 1947 in Chicago, IL, USA. Ph.D. 1977 in physiology from Harvard University.
William R. Kenan, Jr. Professor of Biological Sciences at Columbia University, New York, NY, USA, since 1982.
ROGER Y. TSIEN
Howard Hughes Medical Institute University of California,
San Diego 9500 Gilman Dr, CMM West 310, La Jolla, CA 92093-0647 USA
www.tsienlab.ucsd.edu
US citizen. Born 1952 in New York, NY, USA. Ph.D. in physiology 1977 from Cambridge University, UK.
Professor and Investigator at Howard Hughes Medical Institute, University of California, San Diego, La Jolla, CA, USA since, 1989.
© The Royal Swedish Academy of Sciences
Nobel Prizes and laureates
Six prizes were awarded for achievements that have conferred the greatest benefit to humankind. The 12 laureates' work and discoveries range from proteins' structures and machine learning to fighting for a world free of nuclear weapons.
See them all presented here.
