Advanced information
Scientific background:
The discovery of Hepatitis C virus (pdf)
Scientific background
The discovery of Hepatitis C virus
The 2020 Nobel Prize in Physiology or Medicine is awarded to Harvey J. Alter, Michael Houghton and Charles M. Rice for the discovery of Hepatitis C virus. Hepatitis, from the Greek names for liver and inflammation, is a disease characterized by poor appetite, vomiting, fatigue and jaundice – yellow discoloration of the skin and eyes. Chronic hepatitis leads to liver damage, which may progress to cirrhosis and liver cancer. Viral infection is the leading cause of hepatitis, with some forms persisting without symptoms for many years before life-threatening complications develop. Until the 1960’s, exposure to blood from infected individuals was a major health hazard, with up to 30% risk of chronic hepatitis following surgery or multiple blood transfusions. This risk was only partially reduced by the discovery of the Hepatitis B virus (HBV) and the eventual elimination of HBV-contaminated blood through testing. A more insidious form of hepatitis, characterized by very mild symptoms in the acute phase and a high risk of progression to chronic liver damage and cancer, remained. The work of Alter, Houghton and Rice characterized this form of hepatitis to be a distinct clinical entity, caused by an RNA virus of the Flavivirus family, now known as Hepatitis C virus (HCV). This pioneering work has paved the way for the development of screening methods that have dramatically reduced the risk of acquiring hepatitis from contaminated blood and has led to the development of effective antiviral drugs that have improved the lives of millions of people.
Hepatitis – a serious threat to human health
The first description of hepatitis, dating approximately 400 B.C., is ascribed to the Greek physician Hippocrates, commonly acknowledged as the father of western medicine. He noted the unique course of the disease called “epidemic jaundice” and wrote: “The bile contained in the liver is full of phlegm and blood and erupts. After such an eruption, the patient soon raves, becomes angry, talks nonsense and barks like a dog” [1].
Today, we appreciate that hepatitis may be caused by different types of insults affecting the function and integrity of the liver. Based on causation, the disease may be classified into in-fectious, metabolic, ischemic, autoimmune, genetic or toxic hepatitis, the latter is often associated with alcohol abuse. Infectious hepatitis may be caused by five different types of RNA or DNA viruses that are the most common cause of hepatitis worldwide.
In 1947, long before the discovery of the causative agents, a British hepatologist classified infectious hepatitis into two subtypes: hepatitis A and hepatitis B, based on their different clinical courses and observed routes of transmission [2]. As is currently known, “Infectious or epidemic hepatitis” is caused by RNA viruses of the Picornaviridae (Hepatitis A virus, HAV) or Hepeviridae (Hepatitis E virus, HEV) families. The disease is mainly transmitted through conta-minated food and water, has a short incubation period and manifests as an acute illness that usually resolves and is followed by life-long immunity.
“Serum hepatitis” can be caused by a DNA virus of the Hepadna family (hepatitis B virus, HBV), with or without an associated RNA virus of the Deltaviridae family (Hepatitis D virus, HDV), or an RNA virus of the Flaviviridae family (Hepatitis C virus, HCV) (Figure 1). This form of hepatitis spreads through contact with blood or other bodily fluids and has a long incubation period during which apparently healthy individuals can transmit the disease. In a significant proportion of affected individuals, the infection becomes chronic, which can lead to liver failure and cancer.
The different types of viral hepatitis contribute substantially to the global burden of hepatic diseases. According to the latest WHO Global Hepatitis Report, HAV infection caused 114 million cases of acute hepatitis in 2015, while 257 million people lived with chronic HBV infection and 72 million with chronic HCV infection in the year of the report. Due to their capacity to establish chronic infections, HBV and HCV are major causes of morbidity and mortality with 1.34 million deaths reported in 2015, a 63% increase from 1990, mainly due to HCV infection. The number of deaths is comparable to that caused by tuberculosis (1.5 million deaths reported in 2018) and higher than that caused by AIDS (690,000 deaths reported in 2019) [3].
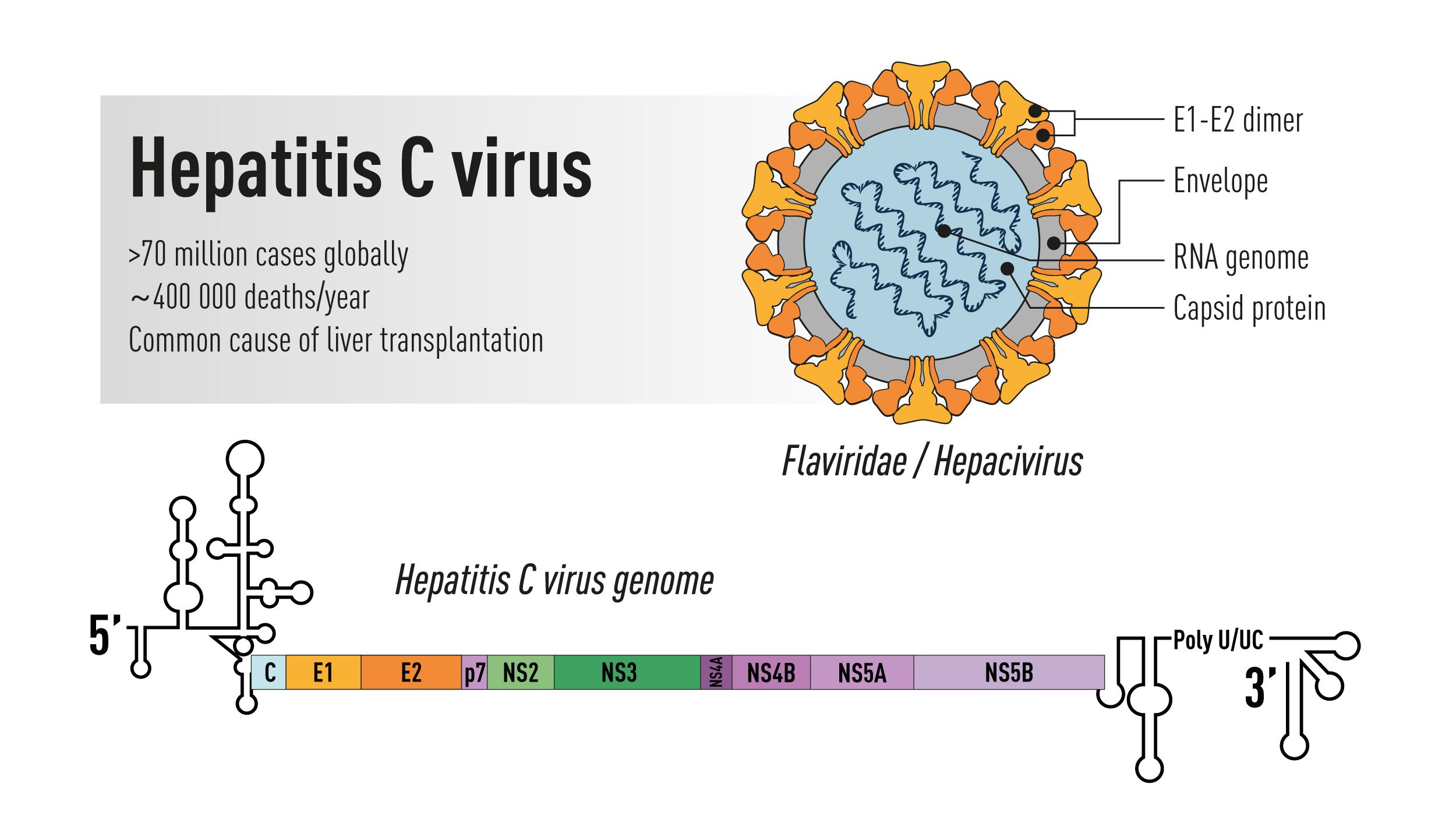
Figure 1. Schematic illustration of the Hepatitis C virus. Top right: the virus particle containing an RNA genome and the viral envelope glycoproteins E1 and E2 exposed on the surface. Bottom: the viral genome encoding a large polyprotein that is cleaved into multiple structural and non-structural proteins with 5’ and 3’ terminal untranslated regions.
The discoveries of the viruses that cause hepatitis are among the most impactful scientific accomplishments of the 20th century. Their identification represents milestone achievements that have revolutionized medicine and sub-stantially improved human health. Through the development of novel technologies, protective vaccines against HAV and HBV are now widely available. The discoveries of HBV and HCV, and the establishment of effective screening routines, have virtually eliminated the risk of transmission via blood products in many parts of the world. Thanks to the development of highly effective drugs against HCV, it is now possible, for the first time in human history, to foresee a future where the threat of this virus infection is substantially reduced and hopefully soon eliminated.
Discovery of the hepatitis B virus
While studying human disease susceptibilities in different populations, Baruch Blumberg, a geneticist working at the National Institutes of Health (NIH) in Bethesda, observed an unusual reaction between the serum of a multiply transfused hemophiliac patient and that of an Australian aborigine; he thought that he had identified a new lipoprotein [4]. Soon thereafter, he could show that the serum from the repeatedly transfused patient detected a new antigen that he called “the Australia-antigen” (Au) [5]. In 1967, the serendipity of a lab technician developing antibodies to the Au-antigen following an episode of jaundice prompted Blumberg to suggest that the Au-antigen may be linked to viral hepatitis. Subsequently, he discovered that the Au antigen found in the serum of individuals affected by post-transfusion hepatitis is part of a virus particle [6]. This enabled the development of immunological tests to screen against this virus and the production of the first vaccine against HBV. Cloning of the virus by Pierre Tiollais, working at the Pasteur Institute in Paris [7], paved the way for the use of genetic engineering techniques for the production of the first highly effective recombinant protein-based vaccine. Before the discovery of HBV, “serum hepatitis” was a serious health threat, with a risk of transmission that could exceed 30% of patients undergoing surgery or receiving contaminated blood products [8]. The development of serological tests that could identify apparently healthy HBV carriers and exclude infected blood resulted in a significant decrease in the frequency of serum hepatitis. For the discovery of HBV, and the development of the first-generation HBV vaccine, Baruch Blumberg was awarded the 1976 Nobel Prize in Physiology or Medicine. However, despite this development, a significant number of individuals still developed chronic hepatitis following blood transfusion, which was a great concern.
Discovery of non-A, non-B hepatitis
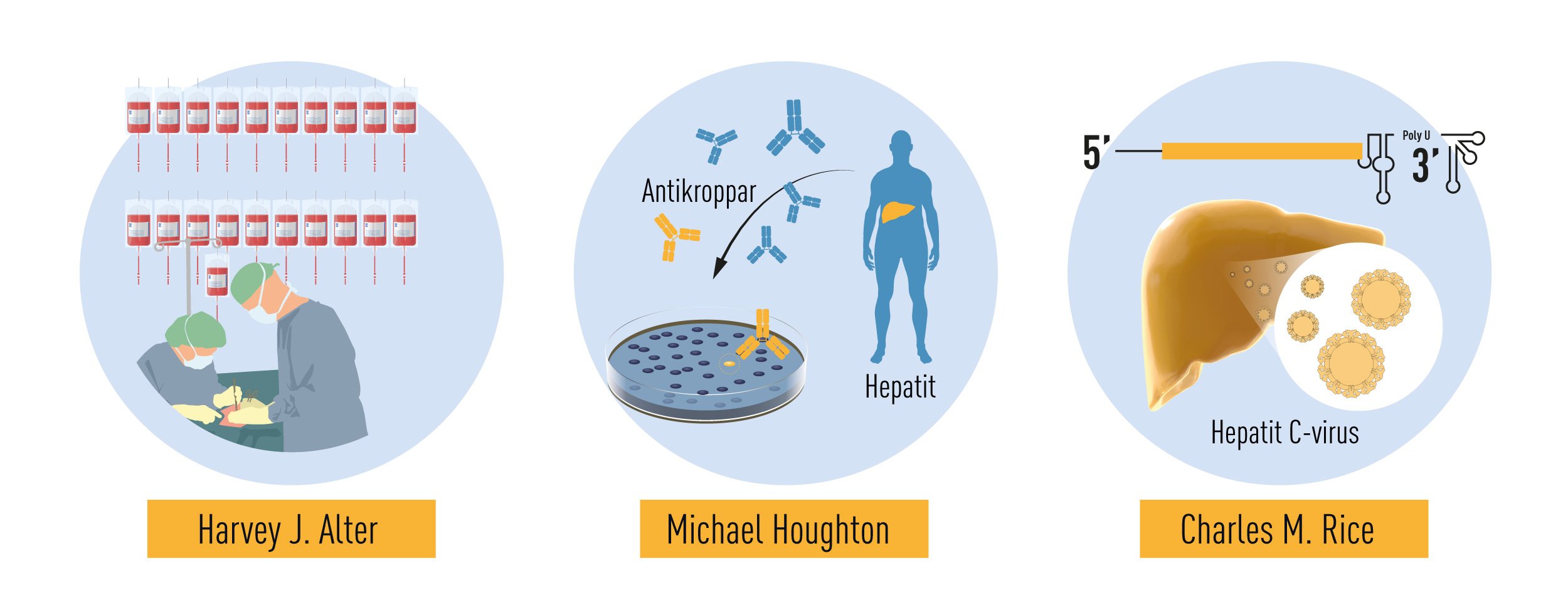
As a young doctor, Harvey J. Alter had collaborated with Baruch Blumberg, making important contributions to the discovery of the Au antigen. He then moved to the NIH Blood Bank where he continued his investigations on post-transfusion hepatitis throughout his career. In the early 1970s, spurred by the discovery of HBV, several research groups began to study the relationship between HBV infection in blood donors and the development of post-transfusion hepatitis [9-13]. It was soon recognized that the exclusion of HBV antibody-positive blood donors prevented only 20% of post-transfusion associated hepatitis. Thus, the remaining 80% of cases appeared to be unrelated to HBV infection. This new form of on “non-B” hepatitis became increasingly prevalent and it differed in the clinical manifestations it caused. While HBV-associated hepatitis had a long incubation period and often presented with severe acute symptoms, the “non-B” serum hepatitis had shorter incubation period and milder symptoms during the acute phase [9, 10]. Among the patients studied by Alter, one individual first developed the mild form of the disease, and later a long-incubation HBV-associated hepatitis [9]. He concluded that at least two different viruses could cause post-transfusion hepatitis.
The initially assumption that the agent responsible for hepatitis A might under certain conditions cause serum hepatitis was abandoned soon after the discovery of the hepatitis A virus. In 1973, two NIH scientists, Stephen Feinstone and Robert H. Purcell, who had just developed the immune-electron microscopy technique, detected a new Picornavirus in the stools of marmosets and patients from acute hepatitis A outbreaks [14]. The virus could be grown in tissue culture, which allowed rapid development of immunological tests for the detection of HAV-specific antibodies in infected patients, and soon thereafter, the production of a protective vaccine. Alter now took advantage of his vast collection of serum samples from non-B hepatitis patients and, in collaboration with Feinstone, Purcell and others, he reported in 1975 that a substantial fraction of non-B hepatitis cases were not caused by HAV or any other known virus [15, 16]. Further epidemiologic evidence suggested that the putative agent(s) responsible for the unexplained hepatitis resembled HBV in terms of the transmission, but led to a chronic infection more frequently [17]. The designation “non-A, non-B hepatitis (NANBH) was coined [15], a working name expected to be short-lived. It was clear that the agent causing NANBH was responsible for an alarming number of post-transfusion hepatitis cases, and the situation was frightening because most of the infected carriers showed no clinical symptoms.
In the following years, investigators failed to make significant progress towards the characterization of the agent responsible for NANBH. In the absence of tests for contaminated blood and tools to identify the causative agent, the disease remained a serious hazard for health workers, patients in need of blood products and intravenous drug abusers. Alter and others partially succeeded in overcoming these challenges in the late 1970’s through the development of a primate model of infection. They showed that serum from patients with acute or chronic NANBH could transmit the disease to chimpanzees [18-20], the only non-human species susceptible to the infection (Figure 2). The availability of an animal model of the disease provided the means to identify morphological changes associated with the infection of hepatocytes and the possibility to characterize the infectious agent with classical virology methods. By determining the infectivity titer of pools of plasma containing the virus, and subjecting aliquots to various treatments, Alter, together with Purcell, established that the putative NANBH virus contained essential lipids [21], a property shared by all enveloped viruses, and had a diameter of approximately 30 to 60 nm [22]. Alter’s pioneering studies had defined a distinct clinical form of post-transfusion hepatitis, trans-mitted by an unknown virus.
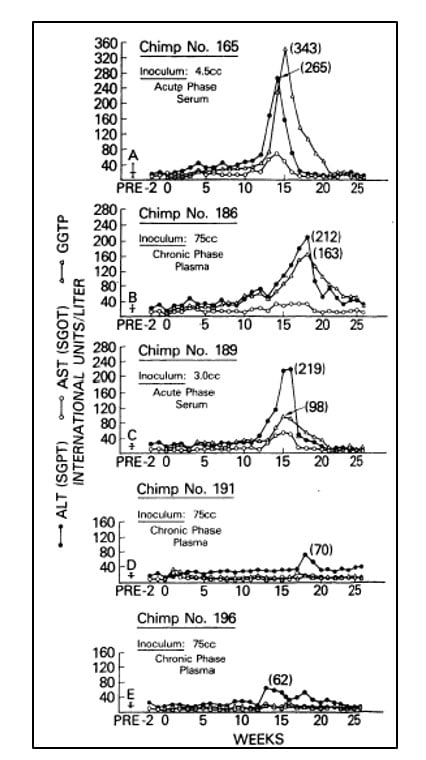
Figure 2. From Alter et al. Lancet 1978. Serial measurements of liver enzymes, alanine aminotransferase (ALT), aspartate aminotrans-ferase (AST) and gamma-glutamyl trans-peptidase (GGTP), in five chimpanzees inoculated with serum or plasma from patients with acute or chronic NANBH.
A new virus
Despite this significant progress, the identity of the virus(es) responsible for NANBH remained frustratingly obscure. The unsuccessful search employing all the traditional methods that had allowed the discovery and characterization of HAV and HBV would continue for more than 10 years. Michael Houghton, working at Chiron Corporation, initiated his hunt for the NANBH virus in 1982 using a molecular approach based on the screening of DNA fragments, also called a complementary DNA (cDNA) library, isolated from infected chimpanzees. The initial screenings identified only genetic material from the host. Attempts to enrich viral sequences by eliminating host sequences that were also found in an uninfected control liver were also unsuccessful. Houghton, then working with Qui-Lim Choo and George Kuo, decided to try a novel immune-screening approach. A cDNA library was generated from RNA isolated from plasma of a NANBH-infected chimpanzees and this was transferred to bacteria using a highly efficient lambda bacteriophage system. Expression of viral antigens was then investigated using serum from a patient with fulminant NANBH, which was presumed to contain antibodies against the unknown virus. Screening one million bacterial colonies using this approach resulted in the identification of one colony that did not contain chimpanzee or human DNA sequences. This was the viral signal they were looking for [23]. The sequence, named clone 5-1-1, hybridized to an RNA of about 10,000 nucleotides. The RNA encoded a large open reading frame (ORF) and exhibited distant homology with the genomes of known RNA viruses. Proteins could be translated from the RNA molecule itself indicating that the virus had a positive strand RNA genome. This allowed classification of the virus, which they named Hepatitis C virus (HCV), as a new member of the Flaviridae family. Further experiments showed that the new viral sequence encoded protein that reacted with sera from a NANBH-infected chimpanzee, but not with sera from control HAV or HBV-infected animals (Figure 3).
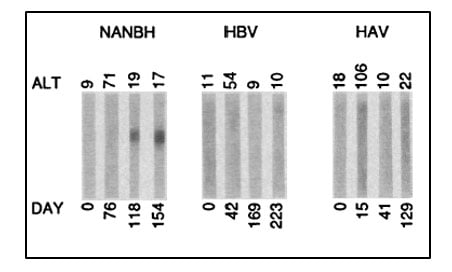
Figure 3. From Choo et al. Science 1989. Immunoblots with sequential serum samples from representative chimpanzees infected with NANBH, HBV or HAV, probed against the protein encoded by the 5-1-1 ORF.
Following identification of the virus, the Houghton team rapidly developed an immunoassay for the detection of HCV-specific antibodies and showed the presence of such antibodies in a blood donor that had transmitted the disease to ten different recipients, and in NANBH patients from Italy, Japan and the USA [24]. These findings esta-blished a firm relationship between infection with the newly discovered HCV and the occurrence of NANBH around the world.
The final proof
The work of Alter and Houghton established a critical link between NANBH and HCV infection. However, it did not constitute a definitive proof of causality because transmission of the disease by transfer of infectious blood could not exclude the involvement of essential co-factors. To conclusively demonstrate causality, the isolation of a virus capable of reproducing the clinical hallmarks of the disease, including chronic liver damage and persistence of infectious virus in the blood of the infected host, was required. A first step towards achieving this goal was made when the groups of Kunitada Shimotohno, working at the National Cancer Center Research Institute in Tokyo, and Charles Rice, working at Washington University in St Louis, in close succession identified a conserved, non-coding region at the 3’ end of the HCV RNA genome, which they surmised could play an important role in virus replication [25-28]. Rice constructed viral RNA genomes containing the conserved 3’ region, injected them into the liver of chimpanzees and looked for evidence of HCV replication but failed to observe newly produced virus in the blood. He then took the next decisive step. Knowing that RNA virus replication is error-prone and that many viral sequences carry inactivating mutations, he engineered a set of RNA genomes that comprised both the conserved 3´ region and a consensus sequence to exclude potential inactivating mutations. He injected the engineered RNA into the liver of chimpanzees and this time the experiment was successful. Productive infection was established, the animals developed clinical signs of hepatitis and infectious virus was found in their blood for several months [29] (Figure 4). A similarly engineered HCV RNA was soon thereafter reported by the laboratory of Jens Bukh [30], also showing that productive infection could be achieved. The work of Charles Rice provided conclusive evidence that HCV alone could cause hepatitis, persist long-term and stimulate a specific antibody response, all features of the human infection.
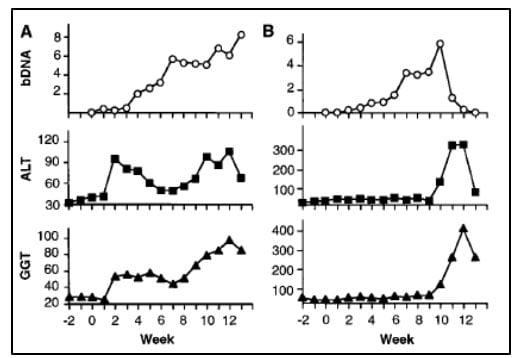
Figure 4. From Kolykhalov et al. Science 1997. HCV viremia and liver enzymes measured weekly in two chimpanzees (shown in A and B, respectively) inoculated with in vitro transcribed RNA encoding the full-length HCV consensus sequence.
New antiviral treatments
The seminal discovery of HCV by this year’s Nobel Laureates paved the way for the development of effective antiviral drugs. While infectious in primates, the full-length clones generated by Rice exhibited poor replicative capacity in cell lines, which hampered in vitro studies of the virus life cycle and testing of candidate antiviral drugs. This obstacle was overcome thanks the work of Ralph Bartenschlager at the University of Heidelberg in Germany who constructed the first HCV sub-genomic clones that replicated with high efficiency in transfected hepatoma cell lines [31]. Further improvement of the technology, and the identification of virus isolates that could replicate in cell lines without the need for adaptive mutations, led to the production of sub-genomic replicons that upon transfection into hepatoma cells resulted in the secretion of virus particles that were infectious [32]. The second obstacle stems from the very restricted host spectrum of HCV – the virus infects only humans and chimpanzees – and the consequent lack of a small animal models for precise assessment of the pathological and immunological profile of the disease, and for pre-clinical testing of candidate drugs. Progress was made when T- and B-cell deficient mice with severe combined immunodeficiency (SCID) could be grafted with human hepatocytes [33] and other models were developed (reviewed in [34]).
The availability of in vitro replication systems and the development of small animal models for in vivo studies facilitated the development of highly effective antiviral drugs that have revolutionized the treatment of HCV infection. Earlier therapeutic regiments, including recombinant type I interferon (IFN) and the nucleoside analog ribavirin, were ineffective and associated with significant side effects. Some improvement was made at the end of the 1990s with the introduction of pegylated-IFN and further improvements were made with the introduction of inhibitors of the NS3/NS4A protease, such a beceprevir, teleprevir and sime-previr. The development of drugs that specifically target the viral RNA-dependent RNA polymerase NS5B, e.g. sofosbuvir [35], and the regulatory replicon protein NS5A, e.g. ledipasvir [36], constituted major breakthroughs in HCV therapeutics. The combination of drugs that target critical viral functions, collectively known as directly acting antivirals (DAAs), proved to be highly effective, caused only minor side effects and strongly diminished the risk of selecting drug resistant variants of the virus [37].
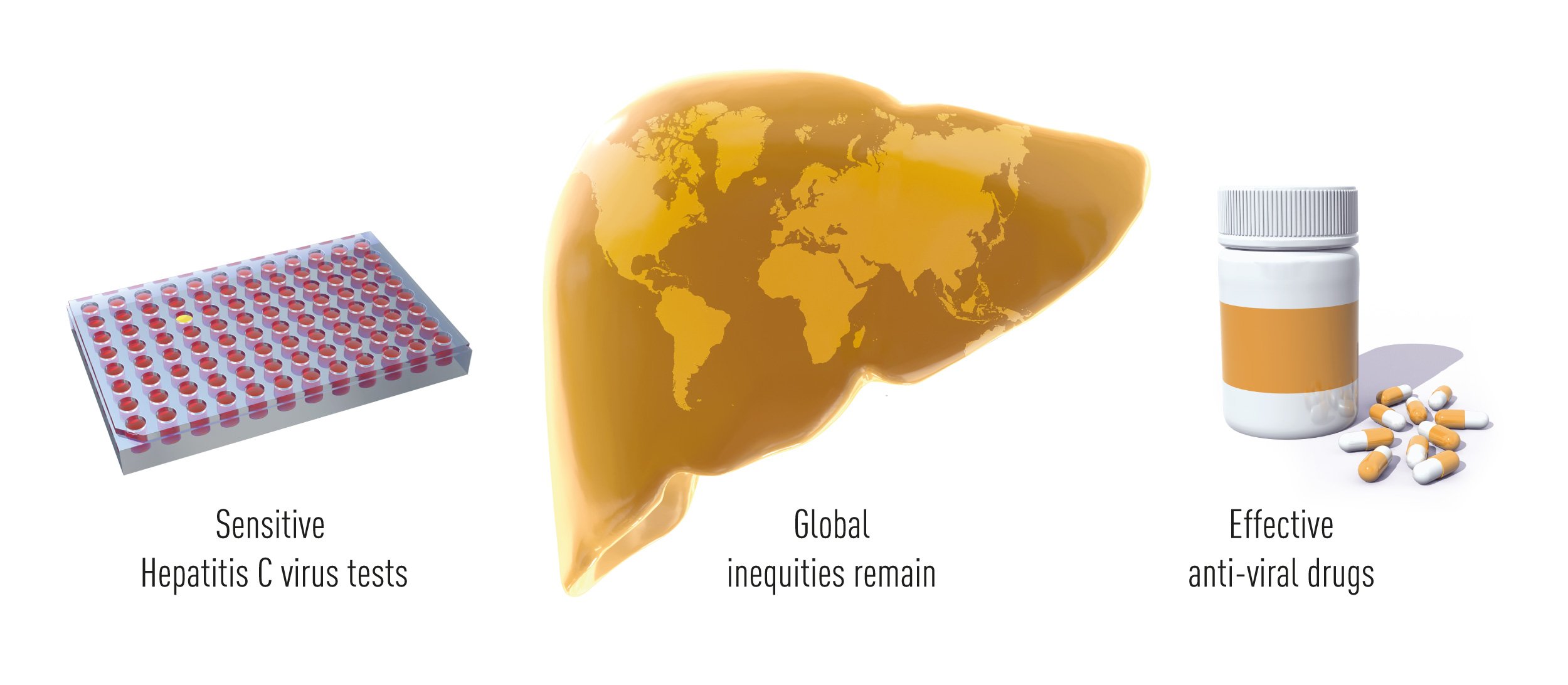
Conclusions
Thanks to the pioneering work of Alter, Houghton and Rice, and many colleagues who built upon their findings, validated tests that identify HCV carriers and allow the elimination of contaminated blood and blood products are broadly available around the world, and effective drugs have changed the fate of HCV infected patients. HCV induced hepatitis is now in many cases a curable disease and the lesions associated with infection are often reversible. Clinical studies have shown that short-term anti-viral treatment cures more than 95% of the patients, including advanced cases who failed to respond to previous therapeutic modalities. This outstanding achieve-ment has already benefitted millions of individuals worldwide. The remaining obstacles towards the eradication of viral hepatitis are now mostly associated with the lack of broad screening campaigns (according the WHO Global Hepatitis Reports 2017 less than 20% of people with HBV or HCV-associated hepatitis have been adequately diagnosed) and with the high cost of the most effective treatments (Figure 5).
References
- Hippocrates.: De Morbus Interna. Œuvres complètes vol 7, London: Publisher Chez J. B. Ballière; 1844: 237-243.
- MacCallum FO: Homologous serum jaundice [An icterus epidemic]. Lancet 1947, 2:691-692.
- Organization WH: Global Hepatitis Report. https://wwwwhoint/hepatitis/publications/global-hepatitis-report2017/en/external 2017.
- Blumberg BS, Alter HJ, Visnich S: A “New” Antigen in Leukemia Sera. JAMA 1965, 191:541-546.
- Blumberg BS, Gerstley BJ, Hungerford DA, London WT, Sutnick AI: A serum antigen (Australia antigen) in Down’s syndrome, leukemia, and hepatitis. Ann Intern Med 1967, 66(5):924-931.
- Bayer ME, Blumberg BS, Werner B: Particles associated with Australia antigen in the sera of patients with leukaemia, Down’s Syndrome and hepatitis. Nature 1968, 218(5146):1057-1059.
- Charnay P, Pourcel C, Louise A, Fritsch A, Tiollais P: Cloning in Escherichia coli and physical structure of hepatitis B virion DNA. Proc Natl Acad Sci U S A 1979, 76(5):2222-2226.
- Chalmers TC, Alter HJ: Management of the Asymptomatic Carrier of the Hepatitis-Associated (Australia) Antigen — Tentative Considerations of the Clinical and Public-Health Aspects. N Engl J Med 1971, 285:613-617.
- Alter HJ, Holland PV, Purcell RH, Lander JJ, Feinstone SM, Morrow AG, Schmidt PJ: Posttransfusion hepatitis after exclusion of commercial and hepatitis-B antigen-positive donors. Ann Intern Med 1972, 77(5):691-699.
- Gocke DJ: A prospective study of posttransfusion hepatitis. The role of Australia Antigen. JAMA 1972, 219(9):1165-1170.
- Grady GF, Bennett AJ, Culhane PO, Forrest JN, Jr., Iber FL: Eight years of surveillance of patients hospitalized with hepatitis: interpretation of data in light of epidemic parenteral drug abuse and availability of testing for hepatitis-associated antigen. J Infect Dis 1972, 126(1):87-91.
- Holland PV, Walsh JH, Morrow AG, Purcell RH: Failure of Australia antibody to prevent post-transfusion hepatitis. Lancet 1969, 2(7620):553-555.
- Hollinger FB, Aach RD, Gitnick GL, Roche JK, Melnick JL: Limitations of solid-phase radioimmunoassay for hb ag in reducing frequency of post-transfusion hepatitis. N Engl J Med 1973, 289(8):385-391.
- Feinstone SM, Kapikian AZ, Purcell RH: Hepatitis A: detection by immune electron microscopy of a viruslike antigen associated with acute illness. Science 1973, 182(4116):1026-1028.
- Alter HJ, Holland PV, Morrow AG, Purcell RH, Feinstone SM, Moritsugu Y: Clinical and serological analysis of transfusion-associated hepatitis. Lancet 1975, 2(7940):838-841.
- Feinstone SM, Kapikian AZ, Purcell RH, Alter HJ, Holland PV: Transfusion-associated hepatitis not due to viral hepatitis type A or B. N Engl J Med 1975, 292(15):767-770.
- Alter HJ, Holland PV, Purcell RH: The emerging pattern of post-transfusion hepatitis. Am J Med Sci 1975, 270(2):329-334.
- Alter HJ, Purcell RH, Holland PV, Popper H: Transmissible agent in non-A, non-B hepatitis. Lancet 1978, 1(8062):459-463.
- Tabor E, Gerety RJ, Drucker JA, Seeff LB, Hoofnagle JH, Jackson DR, April M, Barker LF, Pineda-Tamondong G: Transmission of non-A, non-B hepatitis from man to chimpanzee. Lancet 1978, 1(8062):463-466.
- Hollinger FB, Gitnick GL, Aach RD, Szmuness W, Mosley JW, Stevens CE, Peters RL, Weiner JM, Werch JB, Lander JJ: Non-A, non-B hepatitis transmission in chimpanzees: a project of the transfusion-transmitted viruses study group. Intervirology 1978, 10(1):60-68.
- Feinstone SM, Mihalik KB, Kamimura T, Alter HJ, London WT, Purcell RH: Inactivation of hepatitis B virus and non-A, non-B hepatitis by chloroform. Infect Immun 1983, 41(2):816-821.
- He LF, Alling D, Popkin T, Shapiro M, Alter HJ, Purcell RH: Determining the size of non-A, non-B hepatitis virus by filtration. J Infect Dis 1987, 156(4):636-640.
- Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M: Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 1989, 244(4902):359-362.
- Kuo G, Choo QL, Alter HJ, Gitnick GL, Redeker AG, Purcell RH, Miyamura T, Dienstag JL, Alter MJ, Stevens CE et al: An assay for circulating antibodies to a major etiologic virus of human non-A, non-B hepatitis. Science 1989, 244(4902):362-364.
- Kolykhalov AA, Feinstone SM, Rice CM: Identification of a highly conserved sequence element at the 3′ terminus of hepatitis C virus genome RNA. J Virol 1996, 70(6):3363-3371.
- Tanaka T, Kato N, Cho MJ, Shimotohno K: A novel sequence found at the 3′ terminus of hepatitis C virus genome. Biochem Biophys Res Commun 1995, 215(2):744-749.
- Tanaka T, Kato N, Cho MJ, Sugiyama K, Shimotohno K: Structure of the 3′ terminus of the hepatitis C virus genome. J Virol 1996, 70(5):3307-3312.
- Blight KJ, Rice CM: Secondary structure determination of the conserved 98-base sequence at the 3′ terminus of hepatitis C virus genome RNA. J Virol 1997, 71(10):7345-7352.
- Kolykhalov AA, Agapov EV, Blight KJ, Mihalik K, Feinstone SM, Rice CM: Transmission of hepatitis C by intrahepatic inoculation with transcribed RNA. Science 1997, 277(5325):570-574.
- Yanagi M, Purcell RH, Emerson SU, Bukh J: Transcripts from a single full-length cDNA clone of hepatitis C virus are infectious when directly transfected into the liver of a chimpanzee. Proc Natl Acad Sci U S A 1997, 94(16):8738-8743.
- Lohmann V, Korner F, Koch J, Herian U, Theilmann L, Bartenschlager R: Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 1999, 285(5424):110-113.
- Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Krausslich HG, Mizokami M et al: Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med 2005, 11(7):791-796.
- Mercer DF, Schiller DE, Elliott JF, Douglas DN, Hao C, Rinfret A, Addison WR, Fischer KP, Churchill TA, Lakey JR et al: Hepatitis C virus replication in mice with chimeric human livers. Nat Med 2001, 7(8):927-933.
- Ploss A, Kapoor A: Animal Models of Hepatitis C Virus Infection. Cold Spring Harb Perspect Med 2020, 10(5).
- Bhatia HK, Singh H, Grewal N, Natt NK: Sofosbuvir: A novel treatment option for chronic hepatitis C infection. J Pharmacol Pharmacother 2014, 5(4):278-284.
- Link JO, Taylor JG, Xu L, Mitchell M, Guo H, Liu H, Kato D, Kirschberg T, Sun J, Squires N et al: Discovery of ledipasvir (GS-5885): a potent, once-daily oral NS5A inhibitor for the treatment of hepatitis C virus infection. J Med Chem 2014, 57(5):2033-2046.
- Holmes JA, Rutledge SM, Chung RT: Direct-acting antiviral treatment for hepatitis C. Lancet 2019, 393(10179):1392-1394.
Maria Grazia Masucci, MD PhD, Professor at Karolinska Institutet
Member of the Nobel Assembly
Gunilla Karlsson Hedestam, PhD, Professor at Karolinska Institutet
Member of the Nobel Committee
Illustrations: Mattias Karlén
The Nobel Assembly, consisting of 50 professors at Karolinska Institutet, awards the Nobel Prize in Physiology or Medicine. Its Nobel Committee evaluates the nominations. Since 1901 the Nobel Prize has been awarded to scientists who have made the most important discoveries for the benefit of humankind.
Nobel Prize® is the registered trademark of the Nobel Foundation
Nobel Prizes and laureates
Six prizes were awarded for achievements that have conferred the greatest benefit to humankind. The 14 laureates' work and discoveries range from quantum tunnelling to promoting democratic rights.
See them all presented here.
