Thomas R. Cech
Article

Exploring the new RNA world
by Thomas R. Cech
1989 Nobel Prize laureate in chemistry
This article was published on 3 December 2004.
Not too long ago, most people considered RNA to be just a disposable copy of the really important nucleic acid, DNA. It is the double helix of DNA, after all, that shows up on magazine covers and television; DNA is the material of our genes and chromosomes, the stuff that determines our genetic inheritance. RNA – ribonucleic acid – is a copy of the DNA instructions that serves as a messenger to direct protein synthesis, which is then destroyed after it has fulfilled its function.
My research group in Colorado played a role in discovering novel activities of RNA in the early 1980s. Yale University’s Sidney Altman, who shared the Nobel Prize in Chemistry with me in 1989, made independent discoveries. We both found that RNA could fold into complex shapes and catalyze biochemical reactions, a function previously thought to be restricted to protein enzymes. Thus, RNA was sometimes an active participant in the chemistry of life, not just a passive messenger. We named these RNA enzymes “ribozymes.”
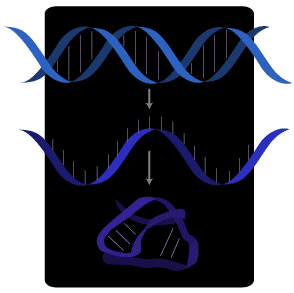
DNA is copied into RNA, which is single-stranded and therefore folds into complex shapes.
An explosion of breakthroughs
Within the past few years, RNA research has reached new heights. It’s now clear that RNA catalysis has a much more central role in biology than many would have guessed. Furthermore, RNA often controls the expression of genes, another role that had been thought to be at least mostly the domain of proteins called “repressors” and “transcription factors.”
An RNA machine makes proteins
One of the most important molecular “machines” in living cells is the ribosome. It translates the message encoded on the mRNA (messenger RNA) to synthesize specific proteins that do most of the work in biology. Proteins can be hormones like insulin or sex hormones; other proteins make muscles contract and hearts beat, and yet other proteins catalyze the digestion of food. Each mRNA encodes a specific protein. The ribosome is remarkable in that it can decode a limitless number of different mRNAs. Think of a videotape: the same tape player can “translate” any videotape into a movie. The tape player is like the ribosome, the tape is the mRNA and the movie is the protein product.
The ribosome is an unusual catalyst, composed of three RNA molecules (four in some species) as well as dozens of proteins. Over the past thirty years there’s been a growing body of evidence that RNA might lie at the catalytic “heart” or active site of the ribosome, with the proteins playing supporting roles. Some of the best evidence has come from the laboratory of Prof. Harry Noller (University of California Santa Cruz, USA). But in biochemistry, “one picture is worth a thousand words,” and obtaining the actual structure of the ribosome was a hugely challenging project despite years of progress by Prof. Ada Yonath (The Weizmann Institute, Rehovot, Israel).
Then, in 2000, the atomic-level picture of the ribosome emerged, showing the complex fold of the RNA molecules buttressed and supported by numerous proteins. One structure showed the site where amino acids are strung together into proteins (the peptidyltransferase center); it is in fact composed of RNA, with no proteins in the vicinity (1). This provides the most direct evidence yet that the ribosome is indeed a ribozyme, an RNA catalyst. Other structures showed how the ribosome is inhibited by antibiotics, how it promotes the interaction of mRNA with transfer RNAs, and how the whole assembly is organized (2, 3, 4).
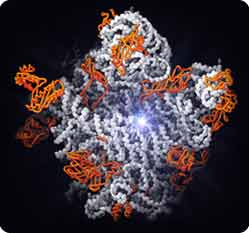
The large subunit of the ribosome. Starburst, active site for protein synthesis. The active site consists of RNA (white strands), not protein (orange), supporting the conclusion that the ribosome is a ribozyme.
Figure by Nenad Ban and Thomas Steitz
Copyright © Howard Hughes Medical Institute
This work may seem esoteric, but in fact it has spurred new approaches to developing antibiotics against pathogenic microbes. After all, protein synthesis is essential for life, and human ribosomes are similar but not identical to microbial ribosomes, so it’s possible to find drugs that inactivate only the latter.
Riboswitches
The switching on and switching off of genes in response to an organism’s needs is one of the most basic of biological control mechanisms. When François Jacob and Jacques Monod were mapping out the genetic regulatory circuit that controls metabolism of a simple sugar (lactose) in a bacterium, they thought for a while that the gene repressor might be RNA. Soon afterwards, however, the lac repressor was isolated – it turned out to be a protein, as were the next hundred or so repressors that were identified. So the idea of an RNA gene-repressor lay dormant for decades.
Very recently, however, this issue has been revisited. In another branch of the bacterial kingdom, RNA elements built into messenger RNAs can directly sense the concentration of small metabolites and turn gene expression on or off in response. These riboswitches fold into intricate structures that can distinguish one metabolite from another (5). Three distinct tricks for switching gene expression have been revealed: the RNA element can cause premature termination of transcription of the mRNA, it can block ribosomes from translating the mRNA, or it can even cleave the mRNA and thereby promote its destruction. This last activity involves an RNA unit directly binding a small-molecule metabolite, which switches the RNA into a conformation that activates its intrinsic self-cleavage activity (6). This “ribozyme riboswitch” represents a new type of biological activity for a catalytic RNA.
RNA interference
The past few years have witnessed the emergence of double-stranded RNA (dsRNA) as a potent regulator of gene expression in higher organisms including humans (7). dsRNA had previously received little attention. Although the DNA of our chromosomes is famously double-stranded, only one strand of the DNA is typically copied into RNA, so the resulting RNA is normally a single strand. (Because it has no partner strand, it folds back onto itself to form a variety of intricate shapes.) dsRNA was mainly studied as a transient intermediate in the replication of RNA viruses, an intermediate that tipped off animal cells to mount an anti-viral response because, after all, double-stranded RNA was “unnatural”.
Now we see that dsRNA isn’t so unnatural after all. Cells maintain a multi-step pathway for dealing with dsRNAs, either endogenous (those made by transcription of their own genes) or exogenous. First, an enzyme called “dicer” cuts the dsRNAs into 20-base pair fragments. One of the two strands is then transferred to a matching sequence on a messenger RNA, and an enzyme called “slicer” then cleaves the mRNA at the position of the duplex (8). The cleaved mRNA is rapidly degraded. In other cellular systems, instead of the mRNA being degraded it stays intact, but the presence of the short RNA duplex renders it somehow untranslatable, so no protein product is made.
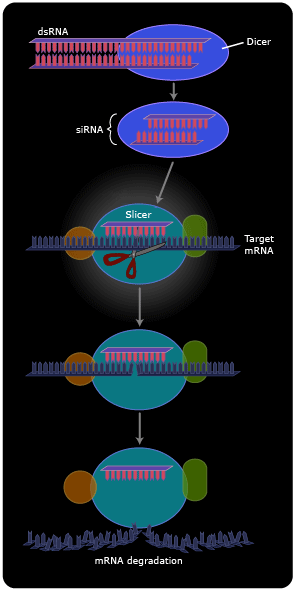
On the one hand, the discovery of RNA interference (RNAi) has led to the identification of many small cellular RNAs that do not encode proteins (and therefore escaped identification in the Human Genome Project) but instead act to regulate the expression of other genes (9). These microRNAs form extensively base-paired “foldback” structures that are then processed by RNAi.
On the other hand, scientists have developed robust new technologies for targeted gene inactivation based on RNAi. They synthesize dsRNA with a sequence that matches the intended target; this dsRNA is often called siRNA (small interfering RNA). They then introduce the dsRNA (or DNA encoding it) into cells, where the RNAi machinery takes over and completes the gene inactivation. Thus, RNAi has become a powerful tool for understanding which genes are important for which biological events. There is currently great excitement concerning the possibility that this research tool could be turned into pharmaceuticals directed against disease-causing genes.
Ribozymes at the chemical level
Coincident with all these new RNA discoveries, great progress is also being made in understanding at the detailed chemical level how the original ribozymes work. Chemists think about chemical reactions in the context of detailed atomic structures of molecules, but for many years catalytic RNAs evaded structural analysis. The RNA molecules simply refused to form well-ordered crystals suitable for x-ray diffraction. This problem was first solved for some of the smaller self-cleaving ribozymes (10, 11), revealing unanticipated intricate folding patterns and the use of RNA bases as proton donors and acceptors to speed the chemical reactions.
Finally this year, the first well-resolved structures of self-splicing introns were solved. The long-anticipated catalytic metal ions can now be directly visualized, held into place by RNA phosphate groups that are in turn positioned by the complex fold of the RNA chain (12, 13, 14). The metal ions in the active site help to stabilize the mid-point or “transition state” of the reaction by neutralizing charge and by orienting the reacting atoms. Exciting crystal structures of portions of the Ribonuclease P ribozyme have also been announced (15). Thus, both subjects of the 1989 Nobel Prize in Chemistry are now seen in atomic detail, making them more amenable to mechanistic analysis.
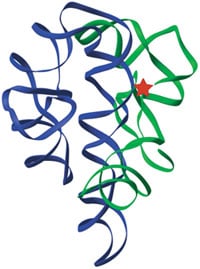
The three-dimensional structure of the original ribozyme, the self-splicing intron of Tetrahymena (13). Green and blue ribbons indicate the path of the RNA backbone in the two major domains of the RNA, and the red star marks the active site.
Figure by Feng Guo
Bibliography
1. Nissen, P., Hansen, J., Ban, N., Moore, P.B. & Steitz, T.A. Science 289, 920-930 (2000).
2. Carter, A.P., Clemons, W.M., Brodersen, D.E., Morgan-Warren, R.J., Wimberly, B. & Ramakrishnan, V. Nature 407, 340-348 (2000).
3. Yusupova, G.Z., Yusupov, M.M., Cate, J.H. & Noller, H.F. Cell 106, 233-241 (2001).
4. Bashan A., Agmon I., Zarivach R., Schluenzen F., Harms J., Berisio R., Bartels H., Franceschi F., Auerbach T., Hansen H.A., Kossoy, E., Kessler M., & Yonath, A. Mol. Cell 11, 91-102 (2003).
5. Batey, R.T., Gilbert, S.D. & Montange, R.K. Nature 432, 411-415 (2004).
6. Winkler, W.C., Nahvi, A., Roth, A., Collins, J.A. & Breaker, R.R. Nature 428, 281-286 (2004).
7. Fire, A., Xu, S., Montgomery, M.K., Kostas, S.A., Driver, S., & Mello, C.C. Nature 391, 806-811 (1998).
8. Song, J.-J., Smith, S.K., Hannon, G.J. & Joshua-Tor, L. Science 305, 1434-1437 (2004).
9. Ruvkun, G. Science 294, 797-799 (2001).
10. Ke, A., Zhou, K., Ding, F., Cate, J.H. & Doudna, J.A. Nature 429, 201-205 (2004).
11. Rupert, P.B. & Ferre-D’Amare, A.R. Nature 410, 761-763 (2001).
12. Adams, P.L., Shahley, M.R., Kosek, A.B., Wang, J. & Strobel, S.A. Nature 430, 45-50 (2004).
13. Guo, F., Gooding, A. & Cech, T.R. Mol. Cell 16, 351-362 (2004).
14. Golden, B., Kim, H. & Chase, E. Nat. Struct. & Mol. Biol., in press (2004).
15. Krasilnikov, A.S., Yang, X., Pan, T. & Mondragon, A. Nature 421, 760-764 (2003).
First published 3 December 2004
Nobel Prizes and laureates
Six prizes were awarded for achievements that have conferred the greatest benefit to humankind. The 12 laureates' work and discoveries range from proteins' structures and machine learning to fighting for a world free of nuclear weapons.
See them all presented here.
