Popular information
Information for the public:
The key to life at the atomic level (pdf)
Populärvetenskaplig information:
Nyckeln till liv på atomnivå (pdf)

The Nobel Prize in Chemistry 2009
At the beginning of the twentieth century, the chemical foundations for life were a mystery. Today we know how many of the most important processes function, all the way down to the atomic level. The 2009 Nobel Prize in Chemistry is awarded for the detailed mapping of the ribosome – the cell’s own protein factory. The ribosome translates the passive DNA information into form and function.
The key to life at the atomic level
The general theory of evolution, which was published by Charles Darwin in 1859, is based on the assumption that an organism’s properties are hereditary and that, every now and then, random changes occur. Successful changes, which increase the chances of survival of the organism in question, are thus carried forward to future generations.
When the scientific community had digested Darwin’s thoughts, some new questions arose: what exactly is being transferred over generations, where do the random changes occur, and how can they manifest themselves in a living organism?
The 2009 Nobel Prize in Chemistry is the third in a series of prizes that show how Darwin’s theories actually function at the level of the atom. Images, generated by means of various X-ray techniques, show how the simple DNA code can manifest itself not only as hearing, feeling and taste, or muscles, bone and skin, but also as thoughts and speech.
The trilogy of prizes began with one of the most famous Nobel Prizes of all, that of 1962, when James Watson, Francis Crick and Maurice Wilkins were recognised for their elaboration of an atomic model of the double-stranded DNA molecule. The second prize in the trilogy was awarded in 2006 to Roger D. Kornberg for X-ray structures that explicate how information is copied to the messenger RNA molecule.
The ribosome translates genetic information into action
The three Nobel Prize Laureates in chemistry for 2009, Ada E. Yonath, Thomas A. Steitz and Venkatraman Ramakrishnan, are rewarded for mapping the ribosome – one of the cell’s most complex machineries – at the atomic level. The ribosome reads the information in messenger RNA, and based upon that information, it produces protein. Scientists refer to this as translation. It is during this translation process, when DNA/RNA language becomes protein language, that life reaches its full complexity.
The body contains tens of thousands of different proteins that control what happens in the body with an astounding precision. Examples of such proteins are: haemoglobin, which transports oxygen from the lungs to the rest of the body; insulin, which controls the sugar level in the blood; antibodies that capture intruding viruses; and keratin, which builds hair and nails.
Ribosomes exist in all cells in all living organisms, from bacteria to human beings. As no living creature can survive without ribosomes, they are the perfect targets for drugs. Many of today’s antibiotics attack the ribosomes of bacteria, but leave those of humans alone. The knowledge that this year’s Nobel Laureates provide us with can thus be of substantial value for the development of new antibiotics. But more about this later on. Let us begin with the mystery that fascinated chemists and biologists during the middle of the twentieth century. How does life function from a chemical point of view?
Proteins – “a string of amino acid pearls”
At the beginning of the 1940s, the mapping of the cell had advanced to such an extent that scientists knew that hereditary traits were carried by chromosomes. Chromosomes consist of nucleic acids (DNA) and proteins (figure 1). The majority of the scientific community thought that the proteins were the carriers of hereditary traits, since they are more complex than DNA.
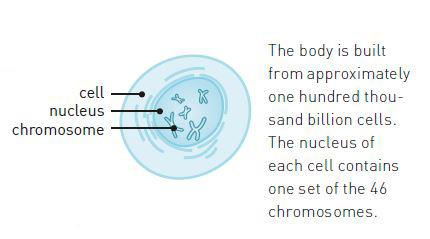
The scientific community was fascinated by the proteins. It was known that some proteins function as building blocks. Others, such as enzymes, trigger and control chemical reactions. However, even though they perform so many different roles in the cell, all proteins consist of the same building blocks, namely twenty different kinds of amino acids. Like pearls on a string, they are linked together in long chains (figure 3). The connection between them, known as a peptide bond, is very stable. A protein chain can consist of anything from ten to tens of thousands of amino acids. Insulin, for example, is much shorter than the haemoglobin of the blood cell.
DNA – too simple a carrier for hereditary traits
The DNA molecule, however, was of little interest to the scientists of the 1940s. In 1871, it was isolated from the cell nucleus by the Swiss scientist Friedrich Miescher. He named the newly discovered molecule nuclein (from the Latin nucleus).
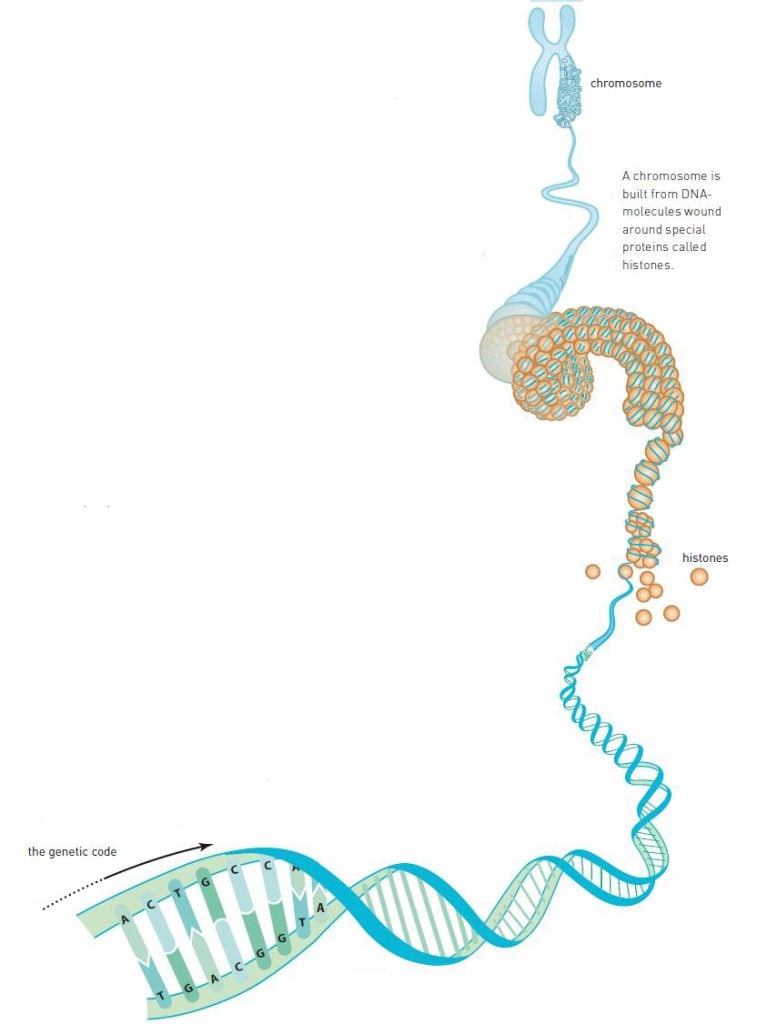
In the same manner as proteins, DNA consists of a string of pearls of smaller molecules. DNA, however, consists of only four different pearls, nucleotides (figure 1). These are carriers of four chemical groups: adenine (A), cytosine (C), guanine (G) and thymine (T).
Four building blocks, however, seemed too few to perform any important task in the cell. Therefore it was believed that DNA mainly functioned as a skeleton for the chromosome’s proteins.
The year of 1944, however, saw the return of the DNA molecule. In what is known as the Avery-MacLeod-McCarty experiment, DNA from dead bacteria was inserted into living bacteria, with the result that the latter were transformed. They changed from being non-virulent to being virulent (triggers of disease).
While the experiment itself was criticized, DNA became a new focus of attention for the scientific community. The understanding of how DNA could carry hereditary traits was first reached with the arrival of an atomic model of the double helix structure.
The elegant double helix
On 28 February 1953, James Watson and Francis Crick at the Cavendish Laboratory at Cambridge University, UK, assembled the pieces of the DNA puzzle. For several years they had tried to understand how the DNA molecule’s four nucleotides could be assembled into a three-dimensional structure.
A clear and sharp X-ray diffraction image, which had been generated by Rosalind Franklin at King’s College in London, showed among other things that DNA forms a spiral, a helix, which consists of two strings. Analyses by the biochemist Erwin Chargaff had shown that DNA, irrespective of what bacteria, insect or animal it comes from, always contains the same amount of A and T and the same amount of C and G.
Watson and Crick, however, had been working with the wrong chemical formulae of the nucleotides. A correction by a colleague led to their understanding that A connects to T and G connects to C (figure 1). The nucleotide pairs, or the base pairs as they are more commonly known, have the same size and fit perfectly in a double helix.
The scientific community then realized that the genetic code is contained within the nucleotide sequences on each of the strands. ATTGCCAT represents something completely different from GCGTATAG. Scientists realized that the sequence of nucleotides controls the sequence of amino acids in proteins. But the question remained: How?
RNA – a cousin of DNA
At the same time as Watson and Crick made their great discovery, the scientific community became more and more interested in another nucleic acid, which primarily is found in the cytoplasm (the part of the cell that is located outside of the nucleus). It had been known for a long time that DNA has a relative, RNA, which also consists of four different nucleotides. Instead of thymine, however, which exists in DNA, the RNA contains uracil (U).
At the beginning of the 1950s, scientists realized that most of the RNA is found in small particles in the cytoplasm (figure 2).
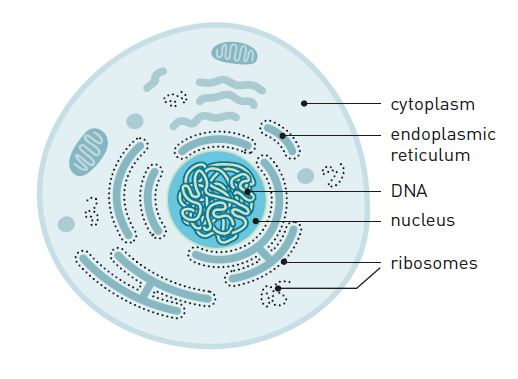
Furthermore, they discovered that this is also the location where proteins are produced. In 1958 they named the protein-producing particle ribosome. It consists of proteins and RNA molecules (ribosomal RNA, or rRNA).
The genetic code was deciphered in the 1960s
Thus, 100 years after Darwin elaborated his theory of evolution, scientists had identified DNA as the molecule that carries hereditary traits. The sequence of nucleotides controls the sequence of amino acids in proteins, which are produced by ribosomes in the cytoplasm. But what was the link between DNA and the ribosomes? They are located on different sides of the nuclear envelope and have no contact with one another (figure 2).
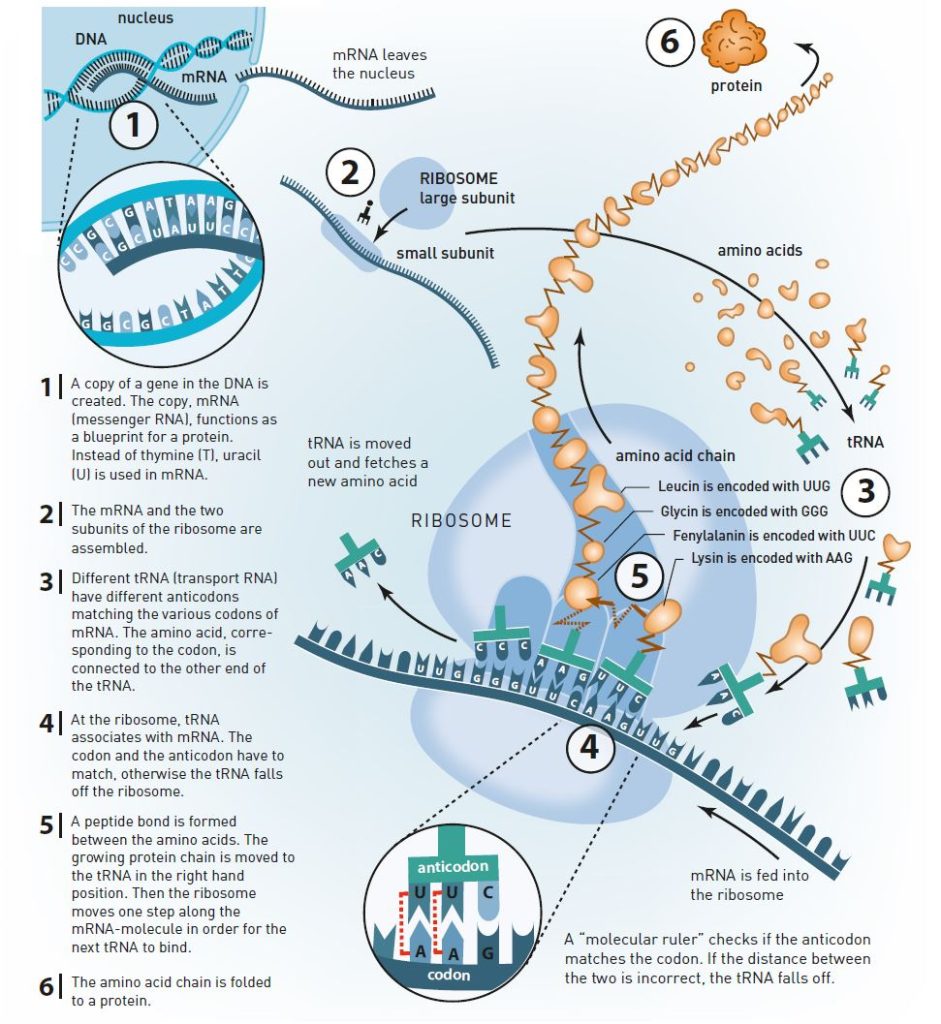
The answer was provided at the beginning of the 1960s. Scientists realized that the genetic message is copied to a RNA molecule (figure 3). They called it messenger RNA (mRNA). mRNA moves outside the nucleus and is caught by the ribosome, which uses mRNA as a blueprint for producing proteins.
When this became known, scientists quickly deciphered the genetic code with the help of artificial mRNA and ribosomes in test tube experiments. The ribosome reads the nucleotides in the mRNA in triplets, or codons. The first codon that became known to the scientists was UUU, which is translated by the ribosome as the amino acid phenylanine. There are 64 different codons and 20 different amino acids, so some of the amino acids are coded by more than one codon.
The reading itself is made by another RNA molecule, transfer RNA (tRNA). At one end of the tRNA, there is an anticodon which is paired with a matching codon on the mRNA molecule in the ribosome (figure 3). At the other end, there is the specific amino acid which matches the codon.
Thus emerged an image of the most fundamental process of life: the manner in which information flows from DNA to RNA and become enzymes and other proteins. The image was still rather schematic, though. As James Watson put it in a review in 1964: “Unfortunately, we cannot accurately describe at the chemical level how a molecule functions unless we know first its structure”. It would take until 2000 before anyone produced a structure that showed how the atoms are located in the ribosome.
Ada Yonath – the strong-willed pioneer
Often a ground-breaking discovery comes from a pioneer who investigates new uncharted territory. In this case, that pioneer was Ada Yonath. At the end of the 1970s, she decided to try to generate X-ray crystallographic structures of the ribosome. At this time, however, most people considered that this was impossible.
In X-ray crystallography, scientists aim X-rays towards a crystal of, for example, a protein (figure 4). When the rays hit the crystal’s atoms they are scattered. On the other side of the crystal, scientists register how the rays have spread out. Previously, this was achieved by using photographic film, which was blackened by the rays. Today one uses CCD detectors, which can be found in digital cameras (and are a focus for the 2009 Nobel Prize in Physics). By analyzing the pattern of dots, scientists can determine exactly how the atoms are positioned in a protein.
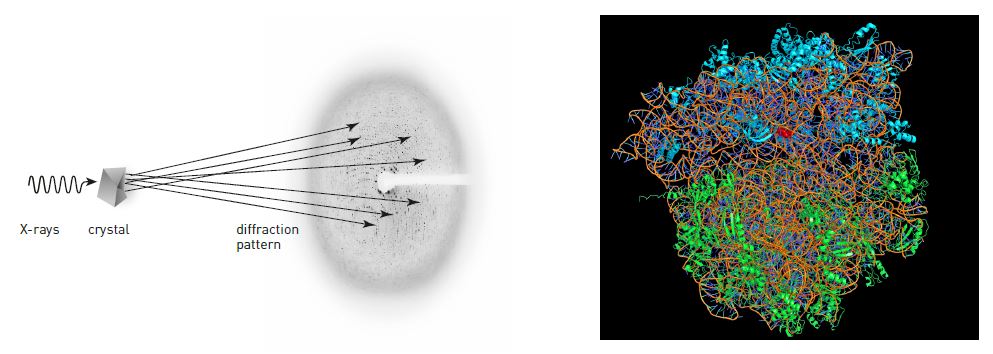
For this to work, however, the crystal has to be almost perfect; the molecules need to form a precise pattern, which is repeated over and over again. With a little luck, when salt water is allowed to slowly evaporate, beautiful salt crystals are formed. However, if a pan full of salt water is allowed to boil dry, the salt only forms a dull layer at the bottom of the pan. Different conditions thus render more or less useful crystals.
To a large extent, this also applies to crystals for X-ray crystallography. To obtain high quality crystals from a protein can be a very tough task, and the larger the protein complex, the harder the task.
Therefore, many people were skeptical of Ada Yonath’s vision. The ribosome is one of the most complicated protein/RNA complexes. It is divided into two parts, “the small subunit” and “the large subunit”. The small subunit in a human ribosome consists of a large RNA molecule and around 32 proteins. The large subunit consists of three RNA molecules and around 46 proteins. Each of the subunits thus consists of thousands of nucleotides and thousands of amino acids, which in turn consist of hundreds of thousands of atoms. Ada Yonath wanted to establish the exact location of each and every one of these atoms in the ribosome.
Warm springs and the Dead Sea – the harder the conditions, the better the crystal
When Ada Yonath decided to crystallize the ribosome, she chose to work with bacteria living under harsh conditions. Geobacillus stearothermophilus can live in warm springs and survives in temperatures up to 75 °C. Ada Yonath’s assumption was that, in order to manage this, the ribosome has to be extremely stable and thus would form better crystals.
In 1980, she had already managed to generate the first three-dimensional crystals of the ribosome’s large subunit. This was a great achievement, although the crystals were far from perfect.
It would actually take another 20 years of hard work before Ada Yonath managed to generate an image of the ribosome where she could determine the location of each atom. She tried many new things. For instance, she stabilized the crystals by freezing them in liquid nitrogen at -196 °C. She also tried to crystallize ribosomes from other resilient micro-organisms. One of them was found very close by – the salt-loving Haloarcula marismortui that lives in the Dead Sea.
Step by step, Ada Yonath got closer to the goal. Eventually, it was realized that the ribosome’s atomic structure could be mapped, and more scientists joined in the race. Among them were Thomas Steitz and Venkatraman Ramakrishnan.
A pattern of millions of black dots packed with meaning
At the beginning of the 1990s, Ada Yonath’s crystals had sufficient quality. The pattern of black dots was detailed enough to determine the location of the atoms in the ribosome crystal. There remained a considerable obstacle, however. It was the “phase problem” of X-ray crystallography. In order to determine a structure from the pattern of black dots, scientists needed to know the “phase angle” for each and every dot. This mathematical information is related to the location of the atoms in the crystal.
A trick frequently employed by scientists in order to determine phase angles, is to soak the crystal in heavy atoms, e.g. mercury. The heavy atoms attach to the surface of the crystal’s ribosomes. By comparing the dotted patterns from crystals with and without heavy atoms, scientists can establish the phase angle.
However, as the ribosomes are so large, too many heavy atoms attached to the ribosome, and it was difficult to immediately determine the phase angle. Additional information was therefore needed in order to solve the phase problem.
It was Thomas Steitz who finally solved the problem. He used images of the ribosome, generated by Joachim Frank, a specialist in electron microscopy. With the help of those images, Thomas Steitz could find out how the ribosomes were oriented and located within the crystal (but the resolution did not allow him to see individual atoms). This information, together with the information from the heavy atoms, finally yielded the phase angle.
Concluding after 20 years of work
In 1998, Thomas Steitz published the first crystal structure of the ribosome’s large subunit. It resembled a dim photograph, and had a resolution of 9 Ångström (one Ångström equals one tenth of a million of a millimetre). It was not possible to see individual atoms, but one could detect the ribosome’s long RNA molecules. This was a decisive breakthrough.
Now that the phase problem had finally been solved, all that remained was to improve the crystals and collect more data, in order to increase the sharpness of the image. This year’s Nobel Laureates reached the finishing line almost simultaneously. In August and September 2000, they published crystal structures with resolutions that allowed interpretation of the atomic locations. Thomas Steitz managed to obtain the structure of the large subunit from Haloarcula marismortui. Ada Yonath and Venkatraman Ramakrishnan obtained the structure of the small subunit from Thermus thermophilus. Thus it was possible to map ribosome functionality at the most basic, atomic level.
The small subunit’s “double checking”
A property of the ribosome, that has fascinated scientists for a long time, is that it seldom makes any errors when it translates DNA/RNA-language into protein language. If an amino acid is incorrectly incorporated, the protein can entirely lose its function, or perhaps even worse, begin to function differently.
For the correct amino acid to be selected depends primarily on the base pairs formed between tRNA and mRNA (figure 3). However, this pairing process is not sufficient to explain the ribosome’s precision.
Venkatraman Ramakrishnan’s crystal structures of the ribosome’s small subunit have been crucial for the understanding of how the ribosome achieves its precision. He identified something that could be described as a molecular ruler (figure 3). Nucleotides in the small subunit’s rRNA measure the distance between the codon in mRNA and the anticodon in tRNA. If the distance is incorrect, the tRNA molecule falls off the ribosome.
Using the ruler twice, the ribosome double-checks that everything is correct. This ensures that errors only occur about once per 100 000 amino acids.
The large subunit places pearls on the string
The role of the large subunit in the ribosome is primarily to synthesize new protein. It triggers the peptide bond formation between the amino acids. To obtain a step-by-step image of the chemical reaction is very difficult, as it occurs at the atomic level and at a daunting speed. In a single ribosome, about 20 peptide bonds can be formed every second.
Thomas Steitz, however, has managed to freeze different moments of the chemical reaction. He has crystallized the large subunit with molecules resembling those that are involved in peptide bond formation. With the help of these structures, scientists have been able to determine which of the ribosome’s atoms are important to the reaction, and how the reaction occurs.
The Laureates of the 2009 Nobel Prize in Chemistry have forged an understanding at the atomic level of how nature can transform something as simple as a four letter code into something as complicated as life itself – just as James Watson predicted in 1964. And research driven by curiosity can also, as so many times before, be of practical use. This time it proves useful in the search for new antibiotics.
The ribosome – a target for new antibiotics
Today, humans have an arsenal of different antibiotics which can be used in the fight against disease-generating bacteria. Many of these antibiotics kill bacteria by blocking the functions of their ribosomes. However, bacteria have become resistant to most of these drugs at an ominous rate. Therefore we need new ones.
This year’s three Nobel Laureates in chemistry have all produced structures that show how different antibiotics bind to the ribosome. Some of them block the tunnel through which the growing proteins leave the ribosome, others prevent the formation of the peptide bond between amino acids. Still others corrupt the translation from DNA/RNA-language into protein language.
Several companies now use the structures of the ribosome in order to develop new antibiotics (figure 5). Some of these are currently undergoing clinical tests, in order to come to grips with the problem of multiresistant bacteria (e.g. MRSA).
The understanding of the ribosome’s structure and function is of great and immediate use to humanity. The discoveries that Ada Yonath, Thomas Steitz and Venkatraman Ramakrishnan have made, are important both for the understanding of how life’s core processes function, and in order to save lives.
Links and further reading
Links
About the ribosome: www.cytochemistry.net/cell-biology/ribosome.htm
About protein chrystallography: www.bio.davidson.edu/Courses/Molbio/MolStudents/spring2003/Kogoy/protein.html
About macromolecular X-ray chrystallography: www-structmed.cimr.cam.ac.uk/Course/Overview/Overview.html
Animations and images: see the Laureates web sites below.
Books
Nierhaus K. H. and Wilson D. N. (2004) Protein synthesis and ribosome structure: translating the genome, WILEY-VCH verlag GmbH&Co. KGaA, Weinheim, 579 p.
Liljas A. (2004) Structural aspects of protein synthesis, World Scientific Publishing Co. Pte. Ltd., Singapore, 308 p.
Walsh C. (2003) Antibiotics: actions, origins, resistance, ASM Press, Washington, 335 p.
The Laureates
VENKATRAMAN RAMAKRISHNAN
Structural Studies Division
MRC Laboratory of Molecular Biology
Hills Road, Cambridge CB2 0QH, England, UK
www.mrc-lmb.cam.ac.uk/ ribo/homepage/ramak/index.html
US citizen. Born in 1952 in Chidambaram, Tamil Nadu, India. Ph.D. in Physics in 1976 from Ohio University, USA.
Senior Scientist and Group Leader at Structural Studies Division, MRC Laboratory of Molecular Biology, Cambridge, UK.
THOMAS A. STEITZ
Molecular Biophysics and Biochemistry Department, Yale University
266 Whitney Avenue, P.O. Box 208114
New Haven, CT 06520-8114 USA
www.mbb.yale.edu/ faculty/pages/steitzt.html
US citizen. Born in 1940 in Milwaukee, WI, USA. Ph.D. in Molecular Biology and Biochemistry in 1966
from Harvard University, USA. Sterling Professor of Molecular Biophysics and Biochemistry
and Howard Hughes Medical Institute Investigator, both at Yale University, CT, USA.
ADA E. YONATH
Department of Structural Biology, Weizmann Institute of Science
76100 Rehovot, Israel
www.weizmann.ac.il/sb/faculty_pages/Yonath/home.html
Israeli citizen. Born in 1939 in Jerusalem, Israel. Ph.D. in X-ray Crystallography in 1968 from the Weizmann Institute of Science, Israel.
Martin S. and Helen Kimmel Professor of Structural Biology and Director of Helen & Milton A. Kimmelman
Center for Biomolecular Structure & Assembly, both at the Weizmann Institute of Science, Rehovot, Israel
© The Royal Swedish Academy of Sciences
Nobel Prizes and laureates
Six prizes were awarded for achievements that have conferred the greatest benefit to humankind. The 12 laureates' work and discoveries range from proteins' structures and machine learning to fighting for a world free of nuclear weapons.
See them all presented here.
