William G. Kaelin Jr
Biographical
 I was born November 23, 1957 in Jamaica, New York City, to William George and Nancy Priscilla (Horn) Kaelin. My mother’s first pregnancy ended in a miscarriage. The obstetrician who delivered me was a friend of my mother’s family. He initially thought I was stillborn and lamented that such obstetrical problems seemed to disproportionately affect his friends. He apparently gave me one last whack, whether for his benefit or mine. Regardless, I began to cry. Eventually, my mother would have four more children, each spaced 2 years apart.
I was born November 23, 1957 in Jamaica, New York City, to William George and Nancy Priscilla (Horn) Kaelin. My mother’s first pregnancy ended in a miscarriage. The obstetrician who delivered me was a friend of my mother’s family. He initially thought I was stillborn and lamented that such obstetrical problems seemed to disproportionately affect his friends. He apparently gave me one last whack, whether for his benefit or mine. Regardless, I began to cry. Eventually, my mother would have four more children, each spaced 2 years apart.
My father went to college and law school at Duke University before becoming an international tax and estate lawyer in Manhattan. My mother majored in mathematics at Adelphi College before becoming an actuary at the Metropolitan Life Insurance Company. She became a homemaker after my birth.
We lived in Jamaica, New York until I was two, at which point we moved to Rockville Centre – a middle-class suburb of New York City on Long Island. There I was enrolled in a public elementary school. I recall the fear of polio and remember being given the oral polio vaccine. That was perhaps my first introduction to the miracles that modern medicine could produce.
On the second day of first grade some male classmates came to my house to play in our small backyard. At one point, while we were roughhousing, I became pinned to the ground under the other boys. Among them was a boy who, I later learned, had been held back in first grade twice for behavioral issues. He was bigger and stronger than the rest of us. He twisted my leg like it was a propeller on a toy airplane (perhaps to emulate the antics of the Three Stooges). I screamed in pain until the boys finally dispersed. I crawled on the ground, unable to walk, until I got to our back door and found my mother. She couldn’t believe that my leg could be broken, so she asked me to stand. I promptly collapsed onto the ground. She drove me to my pediatrician, who confirmed that my leg was broken and put me into a plaster cast to my midthigh. I remember looking at my XRAY on our way home (even I could tell my leg was broken).

Figure 1. William G. Kaelin, Jr. childhood photos, 4 and 11 years old.
For the next 8 weeks I was homebound and largely immobilized, although I could motor around the house if I slid on my buttocks. A tutor came to my house each day so that I wouldn’t fall behind in my classwork. When I returned to school, I was actually so far ahead that I thought I might somehow get in trouble. I remember sitting at my desk and surreptitiously erasing and then redoing afresh the assignments that my classmates were being given for the first time. I sometimes wonder whether all of the individual attention I received while homeschooled contributed to my intellectual development and academic achievements later on. At the same time, I also wonder whether it contributed to my feeling of not quite belonging. This was exacerbated further when I changed schools the next year to attend a Catholic elementary school (St. Agnes). There, I found myself once again immersed among students who already knew one another.
At St. Agnes I was taught by lay women and nuns. This was an era, sadly, when most bright women were told that their career options were: nurse, teacher, or nun. I am sure that some of my female teachers in elementary school (and later high school) would today be lawyers, doctors, college professors, or CEOs. I did extremely well at school until the fifth grade, when two things happened. First, the coolest kid in the class befriended me. By association, I suddenly became cool. Second, I was taught by a young, attractive, nun rather than a lay sexagenarian (at least that’s how old they felt to me). Both of these things led to a lot of acting out on my part, including misbehavior and failure to do my homework. The standard punishments included afterschool detentions and writing out multiple copies of the multiplication table from 1×1 to 12×12. The worse the offense, the more copies you had to write out. I remember once sitting in my backyard writing out 40 copies of the multiplication table. The silver lining was that I was already good in math and now knew multiplication (and hence division) cold.
In those days, I was getting in the low 90s or high 80s (out of 100) in my subjects despite my antics. One day, my fifth grade teacher called my parents and told them that I should be doing much better in school given my aptitude (i.e. I was coasting). After that, my study habits improved somewhat, but they were not exemplary.
My father loved to fish, and we spent many hours fishing together. I think there are many analogies between fishing and science. Two keys to success in fishing are technical expertise (e.g., how to bait a hook) and intuition (e.g., knowing where to fish). Both are easier to learn by apprenticeship than from a book. Similar considerations apply in science. But technical expertise and intuition only get you so far; you also need luck. On any given day the biggest fish might be caught by a novice rather than an expert. The same is true of scientific discovery.
In the 1960s, science was celebrated in the United States due to the cold war and the space race. Like many young boys, I had a toy microscope, chemistry sets (with chemicals that were actually dangerous), electric cars and trains, an erector set, and other toys that stimulated an interest in science. I threw tickertape and confetti at the triumphant Apollo astronauts from my father’s Wall Street office. I also had ample unstructured time to play, whether it was going to the beach, tossing a ball, collecting postage stamps, running a lemonade stand, or making up games with neighborhood children. I regret, however, spending so much time watching television rather than reading.
By the late 1960s, my father could afford a much larger home. He wanted to live in a town within commuting distance of New York that had strong public schools rather than pay five private school tuitions. We moved to Fairfield, Connecticut in the summer of 1970, and I began the eighth grade that September. Entering adolescence is always challenging. In this case, it was complicated by entering a new school system. This exacerbated my shyness. We now had a beautiful house with a swimming pool, but only a handful of other children lived within walking or bicycling distance. Fortunately, I had a few neighborhood friends with whom to play touch football or ice hockey when a nearby pond froze over in winter.
The other problem was that many of my school subjects were taught in tracks, with track one reserved for the most gifted students. As a newcomer, I was placed into track two. This lack of stimulation likely contributed to my poor study habits until and including my high school junior year. I would typically sit in the back of the room and silently scoff at the kids who sat at the front, raising their hands obsequiously at every opportunity. I did the minimum amount of homework necessary and never reviewed the material covered in class that day. My one saving grace was that I did well in mathematics without really trying, and consistently did well on mathematic aptitude tests. As a result, my report cards usually contained an “A” in mathematics and a mixture of “A”s and (mostly) “B”s in other subjects. During this period, it became even clearer that I liked quantitative/objective subjects and disliked qualitative/subjective subjects. I liked learning concepts and ideas and abhorred rote memorization. I was interested in science, but I found biology pretty boring because it was largely descriptive. Chemistry and physics were more my speed. During this time, I learned that I tended to like the things that I was good at and tended to be good at the things that I liked.
During my junior year, my high school, Roger Ludlowe, got a computer terminal that was connected to a mainframe at Fairfield University. Although I was good at mathematics, I did not see a career for myself in mathematics. Computers seemed like an exciting way to put mathematics to work. And I liked that in computer science, as in mathematics, there were objective ways of knowing whether your solutions were correct or “worked”. I began learning various programming languages and would sometimes bicycle to Fairfield University after school to get input from the faculty there.
One day, during my junior year, I was sitting at the Roger Ludlowe computer terminal and saw a lone pamphlet in the waste bin. It described an eight-week National Science Foundation Student Science Training Program in Mathematics and Computer Science held at Florida Atlantic University for 32 “gifted” high school students. I applied and was accepted.
On my flight to Florida, I met another boy on his way to the same program. He had many books he was reading simply for pleasure, including mathematics books and challenging fiction books such as “Gravity’s Rainbow”. He, like all of the other high school students that summer, were amongst the brightest students I had ever met. I was pleasantly surprised, however, to discover that I could hold my own with them in the college level courses that we took that summer. And I discovered that I did better in school when the material was interesting and challenging and when I was surrounded by exceptionally smart people. For the first time in my academic life, I went to the library to checkout extra books to supplement my assigned reading and would work past midnight solving mathematics problems or writing code.

Figure 2. Participants at 1974 National Science Foundation Student Science Training Program at Florida Atlantic University. William G. Kaelin, Jr. is circled.
My fellow summer students and I were given a Russian Math Olympiad test (or its equivalent). It contained the kind of advanced, abstract mathematics problems that could be solved without having had prior formal instruction if you were naturally gifted in mathematics. It was the most challenging test I had ever seen. I was initially crestfallen when I received a score of ~45, the worst grade I had ever gotten. I then looked around and saw that many other students had done much worse. My ego fleetingly recovered until I saw that one of the other students, however, had scored ~145. I shouldn’t have been surprised. He was the one student who was a much better programmer than I was. For example, while I wrote a computer program for playing draw poker as one of my final projects, he wrote a program for playing bridge. I decided that he was the kind of genius who would become an academic mathematician, whereas I better find something more practical. I didn’t see the personal computer revolution coming and didn’t want to write computer code for industrial or military applications. I thought a career in medicine could be the answer (ironically, “Mr. 145” also became a medical doctor).
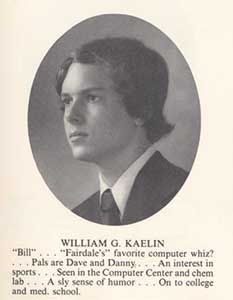
Figure 3. William G. Kaelin, Jr., high school yearbook photo.
Although I was delighted to learn that I wasn’t the dumbest of the 32 students that summer, I certainly had the worst high school grades because of my prior poor study habits. I decided I would try an experiment for my senior year. I would sit in the front of the classroom and actively participate, befriend (rather than belittle) the smart kids in class, do all my homework (and extra credit problems) on time, and review the lessons that we had each day. I successfully petitioned to be placed in all track “1” classes and to take precalculus and advanced placement calculus in parallel rather than in series. I put a bulletin board in my bedroom for my hoped for “A”s. And I determined that I would only have fun on the weekends if had done well at school that week.
My plan worked. I got straight “A”s. I also did very well on the scholastic aptitude test (SAT) and achievement tests required for college. I was a “late bloomer”.
My father, the son of a laborer, was the first member of his family to attend college. Given his parents’ modest means, he sought an up-and-coming school that was still affordable. He applied to Duke, which seemed to fit the bill, and was rejected. He then hitchhiked from his Long Island home to Durham, North Carolina (in the 1950s, this took him several days) to plead his case. The admissions staff, either due to admiration or pity, interviewed him and gave him an aptitude test. His performance earned him admission. While at Duke, my father did a number of odd jobs to pay for his education, including illegally selling sandwiches in the dormitories. He received a scholarship to attend Duke Law School and was allowed to combine his senior year of college with his first year of law school to defray costs. My father was fiercely loyal to Duke and dreamt that I, his eldest son, would also attend Duke.
I applied to Duke and flew to Durham to be interviewed. My father was elated because the associate admissions director, who was one of his former Duke classmates, winked at him as I left the interview to let him know that I would likely be admitted.
I also applied to Harvard and MIT (in those days, “late bloomers” could still get into an elite school. Sadly, this doesn’t seem true today). I visited Harvard with “Mr. 145” and one of his classmates. We stayed overnight in a small dormitory. The 12 students it housed had us stand in the middle of their commons area that evening to tell them our name, our hometown, and our SAT scores. When “Mr. 145” and his friend finished both were told “you will get in”. When I gave my SAT scores, they said “you might want to apply to Brown”. In fairness, my verbal score wasn’t as strong as my math score, and they were correct because I was waitlisted.
I was immensely proud, however, when I was accepted to MIT. I walked into my father’s den and told him I was leaning toward MIT. He put down his scotch and asked, “Don’t you have to be good in math to get into MIT?”, to which I of course said “Yes”. He then said, “I am going to give you a math problem, if you go to MIT, you’ll pay your tuition, but if you go to Duke, I’ll pay your tuition”. I quickly did the math. Truthfully, however, I did go back and forth as to whether Duke might be a better choice for a future physician. And I thought I might get more attention at Duke than at MIT (where I assumed they had a sea of “145”s).
My Duke freshman class contained many students who clearly had functioned at a high level for years. I felt like I was just getting started, and I was fascinated to see what kind of grades I could get if I really applied myself (I am embarrassed that it was still largely about grades for me back then).
I majored in mathematics and, partly to fulfill premedical requirements, chemistry. There were many premedical students at Duke, and it was widely believed that you needed an “A” in Organic Chemistry to get into medical school. I excelled at freshman Inorganic Chemistry because it was logical, conceptual, and quantitative (plus I had an outstanding high school chemistry teacher, Mr. Ralph Minopoli). Organic Chemistry required much more rote memorization. My first Organic Chemistry exam I got a “C” despite studying very hard. The next two exams I got a “B” and then an “A–.” Miraculously, I got an “A+” on the final exam and therefore got an “A” for the term. I had dodged a bullet. I promised myself that I would not dig myself into such a hole the next term, and yet the same test score pattern repeated itself. My assumption is that the rote memorizers couldn’t remember the material from the first exam by the time they got to the final, whereas perhaps I assimilated some concepts after all. However, I haven’t excluded divine intervention.
I earned a higher grade point average at Duke than I did in high school. Ironically, one of my few “Bs” was in Human Physiology, which only reaffirmed my inclination to avoid (largely descriptive) biology courses. I was one of the best mathematics students at Duke, but I worried that I was simply a big fish in a smallish pond. I therefore continued to think about medicine. A friend suggested that I do a laboratory research project to embellish my medical school applications.
My junior year, I met with a physical chemist at Duke about a research project that involved studying the folding of cytochrome C using electron paramagnetic resonance (EPR). I could begin the following summer and continue into the fall semester. He told me that the seven undergraduates who had worked on this project previously had all gone onto medical school. I left his office thinking I had found the golden ticket to medical school.
Now, looking back, I realize I should have asked why my seven predecessors couldn’t bring this project to completion. I soon discovered why. I was given a bench to work at and my predecessors’ notebooks, but no real instructions from my mentor. In fact, he and I barely interacted. None of the other scientists in his laboratory worked on anything even remotely related to my project. Many of them did exclusively dry bench research, and none of them were protein biochemists. I was supposed to covalently couple a spin label to cytochrome C, separate the modified cytochrome C from the unmodified cytochrome C, and then study the labeled Cytochrome by EPR in response to chemical denaturants. I was lost. I assumed that some people intuitively knew what to do in the laboratory and that others, including me, did not. This experience convinced me that I could assimilate old knowledge but lacked what it took to create new knowledge (at least in the laboratory).
That fall, I attended a lecture by a visiting cytochrome C expert. He was kind enough to briefly discuss my project. His first question was simply to ask me “why?” – specifically, “why are you doing this?” He proceeded to tell me that the conditions proposed to covalently add the spin label were already going to denature my protein, and that the purification method recommended by my mentor was going to separate denatured protein from native protein rather than labeled native protein from unlabeled native protein.
I presented my various concerns about my project to my professor. During my senior year Christmas break, I received a special delivery letter indicating that he had given me a “C–” for my independent study project. He also wrote in the margin that “Mr. Kaelin appears to be a bright young man whose future lies outside the laboratory.”

Figure 4. William G. Kaelin, Jr. Duke University Transcript. Note “C–” for laboratory independent study in chemistry.
Despite this debacle, I was accepted to several medical schools, including Duke (I was again rejected by Harvard). The medical school basic science curriculum in those days involved a mind-numbing amount of memorization. One thing that saved me was that Duke compressed the standard two years of basic science into one year. The first year was hard for me, but I felt I could withstand anything for one year (although I did wonder if I had made a mistake not pursuing mathematics and computer science). The other saving grace was that the courses were honors/pass/fail, and it was understood that almost no one got honors. You just plowed through.
In the second year, I took the classical clinical rotations of obstetrics/ gynecology, internal medicine, psychiatry, surgery, and pediatrics. I loved each of my rotations and could now see the knowledge gained in the first year in action. It was exhilarating and reaffirmed my decision to go to medical school. My internal medicine chief resident was Dr. Paul Klotman. I thought he was the smartest, funniest, coolest person I had ever met. I decided then and there that I would be a chief resident one day.
Duke medical students were encouraged to work in a laboratory fulltime for their third year. With some trepidation, I gave laboratory research another try. I worked with Dr. Randy Jirtle, studying tumor blood flow. Randy, in contrast to my first mentor, was very helpful and encouraging. During this time, I read about tumor angiogenesis and the highly vascular tumors linked to von Hippel-Lindau (VHL) disease.

Figure 5. William G. Kaelin. Jr. medical school and housestaff badges.
During my third year, Dr. Michael Bishop came to Duke Medical School to give a lecture, which was exhilarating. In it he described the first oncogenes, some of which encoded kinases. I was sure I was seeing the future of oncology and assumed that this would lead to new therapeutics (e.g., kinase inhibitors). Sadly, over the ensuing years, naysayers would tell me why this was naive. It would take people like Alex Levitzki and Alex Matter to prove them wrong.
Although my second laboratory experience was much better than my first, it wasn’t enough to convince me that I had a future in the laboratory. I did my internship and residency in Internal Medicine at Johns Hopkins. I liked Internal Medicine partly because I liked solving complex clinical puzzles.
My chairman when I was an intern was Dr. Victor McKusick. He taught me the importance of medical history and the power of human genetics. My chairman when I was a resident was Dr. Jack Stobo. He conducted weekly “bench to bedside” chalk talks that introduced me to emerging molecular techniques, such as DNA restriction and Southern blotting. It was pretty clear a revolution was about to happen, even if I didn’t think I would be part of it.
A colleague introduced me to Eric Fearon, who was then a medical student with Dr. Bert Vogelstein. Eric had already published several landmark papers in which he applied modern molecular techniques to gain new insights into cancer genetics. He encouraged me to attend one of Bert’s seminars. At the time, it was believed that solid tumors, in contrast to hematological malignancies, were too heterogeneous and complex to study with modern molecular techniques. Bert’s colon cancer studies shattered that view. This was the second lecture, after Michael Bishop’s lecture, where I knew I was seeing the future.
I loved clinical medicine and was honored to be selected to spend an extra year at Johns Hopkins as one of four assistant chiefs of service (equivalent to chief resident). I learned even more clinical medicine that year and, more importantly, met a beautiful fourth year medical student (although she never reported to me!). Carolyn Scerbo would later become my wife, and the mother of my two beautiful children, Kathryn Grace and Tripp.

Figure 6. Johns Hopkins 1983–1984. Internal Medicine housestaff. William G. Kaelin, Jr., then an intern, is circled.
Chief residents typically love to learn about rare eponymous syndromes, such as von Hippel-Lindau (VHL) Disease. That way, if a trainee challenges their authority on rounds, they can silence them with questions about diseases the trainee has never heard of. For the same reason, chief residents like long differential diagnoses for various signs and symptoms. I had memorized, for example, all of the causes of polycythemia. It struck me as odd that the three hallmark tumors of VHL Disease – kidney cancers, hemangioblastomas, and pheochromocytomas – were among them. This, together with their increased vascularity, suggested to me that the VHL gene was involved in oxygen sensing because angiogenesis and erythropoiesis are normally induced by hypoxia (low oxygen level).
I decided to specialize in medical oncology because I liked multisystem disorders and thought cancer biology was fascinating. I also appreciated that the bane of any internist is distinguishing the sick from the not so sick. This is not an issue with cancer patients.
I moved to Boston in 1987 to be a medical oncology fellow at the Dana-Farber Cancer Institute. I was finally arriving at Harvard, which prompted me to wonder whether Harvard (or I!) had made a mistake. My seven fellow fellows were extremely impressive and most of them had more research experience than I did. I was deemed the most likely of us to become a clinical oncologist rather than a laboratory-based researcher.

Figure 7. William G. Kaelin as Johns Hopkins Internal Medicine Assistant Chief of Service, Circa 1986. Top – With fellow ACSs. Bottom – With his future wife, Carolyn Scerbo, at her medical school graduation in May 1987.
One path to medical oncology board certification involves working in a laboratory for 2 years. I thought it would be exciting to see what life was like in a top laboratory, even if I didn’t expect to succeed myself. I interviewed with Dr. Robert Weinberg at MIT, who had just cloned the retinoblastoma gene RB1. He was clearly receptive to having a physician-scientist in the laboratory, having trained Dr. Steven Friend, who cloned RB1. Bob then got out a ledger and determined that he would have space for me in 3 years. My heart sank because I needed to start that year.
While researching Bob’s laboratory I met Dr. Shelly Bernstein. Shelly, a Weinberg physician-scientist trainee, had just established his own laboratory at the DFCI to study metastasis. I joined Shelly, but I can now appreciate that I was a bit adrift. In my free time, I read a Science paper that described PCR-amplification of a K-Ras exon from a paraffin block. Just for fun, I tried to replicate the experiment. To my astonishment it worked! I remember the exhilaration when I turned on the UV box and saw an ethidium-stained band of the correct size. About 4 months after joining Shelly, he told me that he would be closing his laboratory to return to clinical practice. This seemed like another sign that I was not meant to do science! I was an orphan.
My last attending physician during my clinical oncology year was Dr. David Livingston. I knew he ran a laboratory and I went to him for advice. I showed him my PCR result, which perhaps impressed him. Or maybe he saw something else. In any event, he invited me to join his laboratory. It turned out that he, like Bob Weinberg, was studying the RB1 gene. David’s group and Ed Harlow’s group had discovered that the SV40 T and the adenovirus E1A proteins, respectively, bound to the RB1 gene product pRB. I was to map the T/E1A-binding region of pRB, and the PCR technique I had learned proved useful for rapidly making pRB mutants. The minimal T/E1A-binding region, which we called the pRB “pocket”, turned out to be a hotspot for cancer-associated RB1 mutations. We therefore hypothesized that the pRB pocket might bind to one or more cellular proteins. Our initial strategy to find them was to look for coimmunoprecipitating proteins in anti-pRB immunoprecipitates from cells. The problem, however, was that the supply of our workhorse anti-pRB antibody was unreliable.
David decided we needed to make our own anti-pRB antibodies. I was to express pRB fragments in E.Coli and then inject them into rabbits. The trick was to find a suitable prokaryotic expression vector, since protein expression in E.Coli was still fickle back then.
One day, a company representative left me a brochure for a new prokaryotic expression vector that encoded glutathione S-transferase fused to a protein of interest. Such GST fusions were frequently more soluble than their unfused counterparts and could be purified using glutathione sepharose. Ordinarily, the captured protein would be eluted with glutathione prior to its intended use. I realized, however, that I could express GST fused to the pRB pocket, capture it with glutathione sepharose, and then use the immobilized fusion protein to capture pRB-binding proteins. I eventually found a series of anonymous 35S-labelled proteins that had the properties I was looking for: they bound to wild-type, but not mutant, pRB, and were displaced by T/E1A-derived peptides.
I was thrilled when my findings were published in Cell and when I was allowed to present it at major meetings. After speaking at the 1990 Cold Spring Harbor Symposium, I walked down to the Robertson House pool, where a lone person was enjoying the afternoon. He told me that my talk was wonderful. I told him that his praise was much appreciated because I had just learned that my National Institutes of Health (NIH) K08 physician-scientist fellowship application to support my work had received a very unfavorable score. Indignant, he asked me which study section evaluated my application. When I told him, he said, “that’s outrageous, I chair that study section”. Shortly thereafter, I was notified by the NIH that my grant would be funded after all.
I still didn’t know the identities of my pRB-binding proteins, or which of them bound directly to pRB. My colleague Myles Brown then told me about the work of Michael Blanar, who had radiolabeled a recombinant protein using radioactive ATP, heart muscle kinase, and a genetically encoded phosphoacceptor site. I redesigned my GST fusion vector so that this phosphoacceptor site was encoded between the GST and pRB moieties. After considerable troubleshooting, I finally produced biologically active, radiolabeled GST-pRB, which I used to probe whole cell extracts in “far western blot” assays. I knew the system worked because I could detect E1A and T in appropriate cell extracts. I also saw a doublet of ~50 kD, which prompted me to team up with one of Dr. Sam Speck’s postdoctoral fellows, Erik Flemington. Erik helped me screen a lambda phage expression library with my radiolabeled GST-pRB. David Livingston, Joe Nevins, and Pradid Raychaudhuri had reported that pRB could bind (directly or indirectly) to a DNA-binding activity called “E2F”. I was elated when my expression cloning strategy yielded the first E2F cDNA (coined E2F1).
Why was I so successful as a postdoctoral fellow? In stark contrast to my first laboratory experience, I was given a great project to work on, in a great laboratory, and with a great mentor. My expectations of myself were so low that I was exhilarated when even the most prosaic experiment, such as restricting DNA or transforming bacteria, actually worked! Importantly, I was still relatively naïve, which can be a blessing in the laboratory. I ambitiously tried many experiments that a more jaded scientist would dismiss as being too risky. And I didn’t know the usual solutions for many problems. This forced me to be resourceful and to look at problems afresh.
In 1992, I started my own laboratory down the hallway from David’s laboratory. I initially did a few E2F experiments, but it was obvious that there were many good laboratories ready to work on this protein. I didn’t want to look back on my scientific career and realize that everything I had done would have been done with or without me. That would hardly justify leaving clinical medicine, which I loved. Moreover, people wisely advised me to begin differentiating my science from David’s science. I briefly attempted to expression clone the ATM gene using a functional complementation assay, but without success. In the summer of 1993, however, I read a Science paper describing the cloning of the VHL tumor suppressor gene and decided that it would be my focus. I assumed that studying the VHL gene would teach us about the pathogenesis of kidney cancer (which is one of the ten most common cancers), about the control of angiogenesis, and potentially about oxygen sensing. This work is described more fully in my written Nobel Lecture. It was supported by the NIH, the Howard Hughes Medical Institute (which I joined in 1998), the Doris Duke Charitable Foundation, the Murray Foundation, and the late Dr. William Shelton.
As a young faculty member, I served as an inpatient clinical attending physician one month a year and would also “moonlight” several nights a month in a community hospital intensive care unit. I stopped seeing patients, however, by the late 1990s. Firstly, I never wanted people to say I was a good scientist for a physician-scientist. I tell young physicians that if their choice is to be an “A” clinician or a “B” scientist, they should choose the former. If I was going to try to be an “A” scientist, I knew I had to do it full-time. Second, I soon realized that most of the cancer patients referred to the DFCI had already been diagnosed. The diagnostic puzzles I was solving often related to the iatrogenic complications of their treatments. There is also a saying on the wards that common things are common. There are many things in medicine that are interesting the first or second time you see them, but much less interesting the 20th time you see them. Solving puzzles in the laboratory began to give me the joy I had once found seeing patients. It was also painfully clear that the therapies available for most of my cancer patients were woefully inadequate. I thought that more effective therapies would require the kind of deep insights into cancer pathogenesis that modern molecular biology offered. Lastly, you can’t, by definition, devote yourself fully to two careers. When I was younger, I believed that my patients were lucky to have me as a doctor and I promised myself that I would stop seeing patients if I could no longer defend that view. I had too much respect for my full-time clinical colleagues and my patients to become a part-time clinician with waning skills.
In 2003, several years after the work that led to my Nobel Prize, my wife Carolyn, herself a beloved breast cancer surgeon, developed breast cancer. She ultimately required a mastectomy and chemotherapy. Even though I had taken care of hundreds of cancer patients and their families, I now had a much more visceral understanding of their anguish and pain. Carolyn survived her breast cancer only to develop an unrelated glioblastoma in 2010. She underwent two major brain operations, each requiring months of rehabilitation in order for her to walk again, as well as radiation therapy and chemotherapy. We assembled a “dream team” of talented clinicians and scientists who helped us. For example, her tumor’s genome was sequenced early on as a guide to specific targeted agents. Ultimately, however, we lost her in 2015. Carolyn and I would often joke about what it would be like if I ever won the Nobel Prize. It was bittersweet to receive the Prize without her at my side.
This autobiography/biography was written at the time of the award and later published in the book series Les Prix Nobel/ Nobel Lectures/The Nobel Prizes. The information is sometimes updated with an addendum submitted by the Laureate.
Nobel Prizes and laureates
See them all presented here.
