Advanced information
Scientific background:
Discoveries of mechanisms for autophagy (pdf)

Scientific background
Discoveries of Mechanisms for Autophagy
The 2016 Nobel Prize in Physiology or Medicine is awarded to Yoshinori Ohsumi for his discoveries of mechanisms for autophagy. Macroautophagy (“self-eating”, hereafter referred to as autophagy) is an evolutionarily conserved process whereby the eukaryotic cell can recycle part of its own content by sequestering a portion of the cytoplasm in a double-membrane vesicle that is delivered to the lysosome for digestion. Unlike other cellular degradation machineries, autophagy removes long-lived proteins, large macro-molecular complexes and organelles that have become obsolete or damaged. Autophagy mediates the digestion and recycling of non-essential parts of the cell during starvation and participates in a variety of physiological processes where cellular components must be removed to leave space for new ones. In addition, autophagy is a key cellular process capable of clearing invading microorganisms and toxic protein aggregates, and therefore plays an important role during infection, in ageing and in the pathogenesis of many human diseases. Although autophagy was recognized already in the 1960’s, the mechanism and physiological relevance remained poorly understood for decades. The work of Yoshinori Ohsumi dramatically transformed the understanding of this vital cellular process. In 1993, Ohsumi published his seminal discovery of 15 genes of key importance for autophagy in budding yeast. In a series of elegant subsequent studies, he cloned several of these genes in yeast and mammalian cells and elucidated the function of the encoded proteins. Based on Yoshinori Ohsumi’s seminal discoveries, the importance of autophagy in human physiology and disease is now appreciated.
The mystery of autophagy
In the early 1950’s, Christian de Duve was interested in the action of insulin and studied the intracellular localization of glucose-6-phosphatase using cell fractionation methods developed by Albert Claude. In a control experiment, he also followed the distribution of acid phosphatase, but failed to detect any enzymatic activity in freshly isolated liver fractions. Remarkably, the enzymatic activity reappeared if the fractions were stored for five days in a refrigerator1. It soon became clear that proteolytic enzymes were sequestered within a previously unknown membrane structure that de Duve named the lysosome1,2. Comparative electron microscopy of purified lysosome-rich liver fractions and sectioned liver identified the lysosome as a distinct cellular organelle3. Christian de Duve and Albert Claude, together with George Palade, were awarded the 1974 Nobel Prize in Physiology or Medicine for their discoveries concerning the structure and functional organization of the cell.
Soon after the discovery of the lysosome, researchers found that portions of the cytoplasm are sequestered into membranous structures during normal kidney development in the mouse4. Similar structures containing a small amount of cytoplasm and mitochondria were observed in the proximal tubule cells of rat kidney during hydronephrosis5. The vacuoles were found to colocalize with acid-phosphatase-containing granules during the early stages of degeneration and the structures were shown to increase as degeneration progressed5. Membrane structures containing degenerating cytoplasm were also present in normal rat liver cells and their abundance increased dramatically following glucagon perfusion6 or exposure to toxic agents7.
Recognizing that the structures had the capacity to digest parts of the intracellular content, Christian de Duve coined the term autophagy in 1963, and extensively discussed this concept in a review article published a few years later8. At that time, a compelling case for the existence of autophagy in mammalian cells was made based on results from electron microscopy studies8. Autophagy was known to occur at a low basal level, and to increase during differentiation and remodeling in a variety of tissues, including brain, intestine, kidney, lung, liver, prostate, skin and thyroid gland4,7-13. It was speculated that autophagy might be a mechanism for coping with metabolic stress in response to starvation6 and that it might have roles in the pathogenesis of disease5. Furthermore, autophagy was shown to occur in a wide range of single cell eukaryotes and metazoa, e.g. amoeba, Euglena gracilis, Tetrahymena, insects and frogs8,14, pointing to a function conserved throughout evolution.
During the following decades, advances in the field were limited. Nutrients and hormones were reported to influence autophagy; amino acid deprivation induced15, and insulin-stimulation suppressed16 autophagy in mammalian tissues. A small molecule, 3-methyladenine, was shown to inhibit autophagy17. One study using a combination of cell fractionation, autoradiography and electron microscopy provided evidence that the early stage of autophagy included the formation of a doublemembrane structure, the phagophore, that extended around a portion of the cytoplasm and closed into a vesicle lacking hydrolytic enzymes, the autophagosome18 (Figure 1).
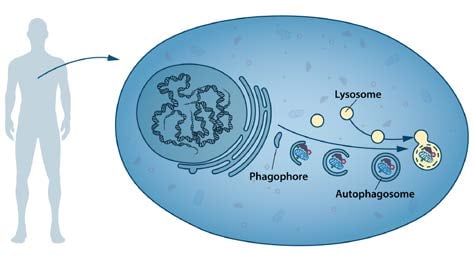
Despite many indications that autophagy could be an important cellular process, its mechanism and regulation were not understood. Only a handful of laboratories were working on the problem, mainly using correlative or descriptive approaches and focusing on the late stages of autophagy, i.e. the steps just before or after fusion with the lysosome. We now know that the autophagosome is transient and only exists for ~10-20 minutes before fusing with the lysosome, making morphological and biochemical studies very difficult.
In the early 1990’s, almost 30 years after de Duve coined the term autophagy, the process remained a biological enigma. Molecular markers were not available and components of the autophagy machinery were elusive. Many fundamental questions remained unanswered: How was the autophagy process initiated? How was the autophagosome formed? How important was autophagy for cellular and organismal survival? Did autophagy have any role in human disease?
Discovery of the autophagy machinery
In the early 1990’s Yoshinori Ohsumi, then an Assistant Professor at Tokyo University, decided to study autophagy using the budding yeast Saccharomyces cerevisae as a model system. The first question he addressed was whether autophagy exists in this unicellular organism. The yeast vacuole is the functional equivalent of the mammalian lysosome. Ohsumi reasoned that, if autophagy existed in yeast, inhibition of vacuolar enzymes would result in the accumulation of engulfed cytoplasmic components in the vacuole. To test this hypothesis, he developed yeast strains that lacked the vacuolar proteases proteinase A, proteinase B and carboxy-peptidase19. He found that autophagic bodies accumulated in the vacuole when the engineered yeast were grown in nutrientdeprived medium19, producing an abnormal vacuole that was visible under a light microscope. He had now identified a unique phenotype that could be used to discover genes that control the induction of autophagy. By inducing random mutations in yeast cells lacking vacuolar proteases, Ohsumi identified the first mutant that could not accumulate autophagic bodies in the vacuole20; he named this gene autophagy 1 (APG1). He then found that the APG1 mutant lost viability much quicker than wild-type yeast cells in nitrogendeprived medium. As a second screen he used this more convenient phenotype and additional characterization to identify 75 recessive mutants that could be categorized into different complementation groups. In an article published in FEBS Letters in 1993, Ohsumi reported his discovery of as many as 15 genes that are essential for the activation of autophagy in eukaryotic cells20. He named the genes APG1-15. As new autophagy genes were identified in yeast and other species, a unified system of gene nomenclature using the ATG abbreviation was adopted21. This nomenclature will be used henceforth in the text.
During the following years, Ohsumi cloned several ATG genes22-24 and characterized the function of their protein products. Cloning of the ATG1 gene revealed that it encodes a serine/threonine kinase, demonstrating a role for protein phosphorylation in autophagy24. Additional studies showed that Atg1 forms a complex with the product of the ATG13 gene, and that this interaction is regulated by the target of rapamycin (TOR) kinase23,25. TOR is active in cells grown under nutrient-rich conditions and hyper-phosphorylates Atg13, which prevents the formation of the Atg13:Atg1 complex. Conversely, when TOR is inactivated by starvation, dephosphorylated Atg13 binds Atg1 and autophagy is activated25. Subsequently, the active kinase was shown to be a pentameric complex26 that includes, in addition to Atg1 and Atg13, Atg17, Atg29 and Atg31. The assembly of this complex is a first step in a cascade of events needed for formation of the autophagosome.
The formation of the autophagosome involves the integral membrane protein Atg9, as well as a phosphatidylinositol-3 kinase (PI3K) complex26 composed of vacuolar protein sorting-associated protein 34 (Vps34), Vps15, Atg6, and Atg14. This complex generates phosphatidylinositol-3 phosphate and additional Atg proteins are recruited to the membrane of the phagophore. Extension of the phagophore to form the mature autophagosome involves two ubiquitin-like protein conjugation cascades (Figure 2).
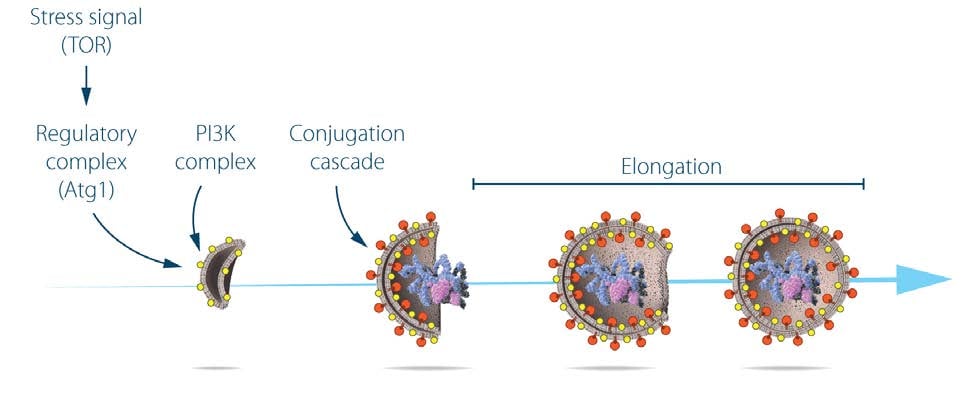
Studies on the localization of Atg8 showed that, while the protein was evenly distributed throughout the cytoplasm of growing yeast cells, in starved cells, Atg8 formed large aggregates that colocalized with autophagosomes and autophagic bodies27. Ohsumi made the surprising discovery that the membrane localization of Atg8 is dependent on two ubiquitin-like conjugation systems that act sequentially to promote the covalent binding of Atg8 to the membrane lipid phosphatidylethanolamine. The two systems share the same activating enzyme, Atg7. In the first conjugation event, Atg12 is activated by forming a thioester bond with a cysteine residue of Atg7, and then transferred to the conjugating enzyme Atg10 that catalyzes its covalent binding to the Atg5 protein26,28,29. Further work showed that the Atg12:Atg5 conjugate recruits Atg16 to form a trimolecular complex that plays an essential role in autophagy by acting as the ligase of the second ubiquitin-like conjugation system30. In this second unique reaction, the C-terminal arginine of Atg8 is removed by Atg4, and mature Atg8 is subsequently activated by Atg7 for transfer to the Atg3 conjugating enzyme31. Finally, the two conjugation systems converge as the Atg12:Atg5:Atg16 ligase promotes the conjugation of Atg8 to phosphatidylethanolamine26,32. Lipidated Atg8 is a key driver of autophagosome elongation and fusion33,34. The two conjugation systems are highly conserved between yeast and mammals. A fluorescently tagged version of the mammalian homologue of yeast Atg8, called light chain 3 (LC3), is extensively used as a marker of autophagosome formation in mammalian systems35, 36.
Ohsumi and colleagues were the first to identify mammalian homologues of the yeast ATG genes, which allowed studies on the function of autophagy in higher eukaryotes. Soon after, genetic studies revealed that mice lacking the Atg5 gene are apparently normal at birth, but die during the first day of life due to inability to cope with the starvation that precedes feeding37. Studies of knockout mouse models lacking different components of the autophagy machinery have confirmed the importance of the process in a variety of mammalian tissues26,38.
The pioneering studies by Ohsumi generated an enormous interest in autophagy. The field has become one of the most intensely studied areas of biomedical research, with a remarkable increase in the number of publications since the early 2000’s.
Different types of autophagy
Following the seminal discoveries of Ohsumi, different subtypes of autophagy can now be distinguished depending on the cargo that is degraded. The most extensively studied form of autophagy, macroautophagy, degrades large portions of the cytoplasm and cellular organelles. Non-selective autophagy occurs continuously, and is efficiently induced in response to stress, e.g. starvation. In addition, the selective autophagy of specific classes of substrates – protein aggregates, cytoplasmic organelles or invading viruses and bacteria – involves specific adaptors that recognize the cargo and targets it to Atg8/LC3 on the autophagosomal membrane39. Other forms of autophagy include microautophagy40, which involves the direct engulfment of cytoplasmic material via inward folding of the lysosomal membrane, and chaperone-mediated autophagy (CMA). In CMA, proteins with specific recognition signals are directly translocated into the lysosome via binding to a chaperone complex41.
Autophagy in health and disease
Insights provided by the molecular characterization of autophagy have been instrumental in advancing the understanding of this process and its involvement in cell physiology and a variety of pathological states (Figure 3). Autophagy was initially recognized as a cellular response to stress, but we now know that the system operates continuously at basal levels. Unlike the ubiquitinproteasome system that preferentially degrades short-lived proteins, autophagy removes long-lived proteins and is the only process capable of destroying whole organelles, such as mitochondria, peroxisomes and the endoplasmic reticulum. Thus, autophagy plays an essential role in the maintenance of cellular homeostasis. Moreover, autophagy participates in a variety of physiological processes, such as cell differentiation and embryogenesis that require the disposal of large portions of the cytoplasm. The rapid induction of autophagy in response to different types of stress underlies its cytoprotective function and the capacity to counteract cell injury and many diseases associated with ageing.
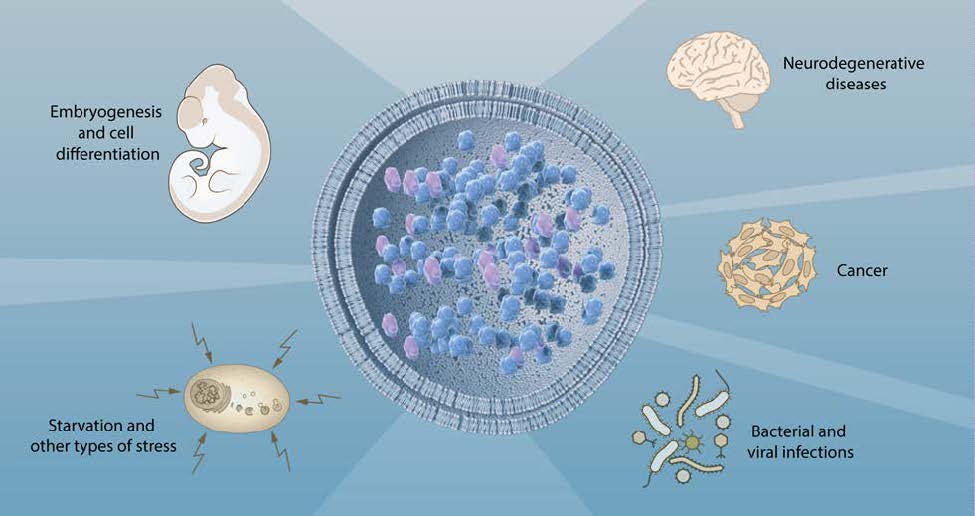
Because the deregulation of the autophagic flux is directly or indirectly involved in a broad spectrum of human diseases, autophagy is a particularly interesting target for therapeutic intervention. An important first insight into the role of autophagy in disease came from the observation that Beclin-1, the product of the BECN1 gene, is mutated in a large proportion of human breast and ovarian cancers. BECN1 is a homolog of yeast ATG6 that regulates steps in the initiation of autophagy42. This finding generated substantial interest in the role of autophagy in cancer43.
Misfolded proteins tend to form insoluble aggregates that are toxic to cells. To cope with this problem the cell depends on autophagy44. In fly and mouse models of neurodegenerative diseases, the activation of autophagy by inhibition of TOR kinase reduces the toxicity of protein aggregates45. Moreover, loss of autophagy in the mouse brain by the tissue-specific disruption of Atg5 and Atg7 causes neurodegeneration46,47. Several autosomal recessive human diseases with impaired autophagy are characterized by brain malformations, developmental delay, intellectual disability, epilepsy, movement disorders and neurodegeneration48.
The capacity of autophagy to eliminate invading microorganisms, a phenomenon called xenophagy, underlies its key role in the activation of immune responses and the control of infectious diseases49,50. Viruses and intracellular bacteria have developed sophisticated strategies to circumvent this cellular defense. Additionally, microorganisms can exploit autophagy to sustain their own growth.
Conclusion
The discovery of autophagy genes, and the elucidation of the molecular machinery for autophagy by Yoshinori Ohsumi have led to a new paradigm in the understanding of how the cell recycles its contents. Because of his pioneering work, autophagy is recognized as a fundamental process in cell physiology with major implications for human health and disease.
Nils-Göran Larsson and Maria G. Masucci Karolinska Institutet
References
1. de Duve, C. (2005). The lysosome turns fifty. Nat Cell Biol 7, 847–849.
2. de Duve, C., Pressman, B.C., Gianetto, R., Wattiaux, R., and Appelmans, F. (1955) Tissue fractionation studies. 6. Intracellular distribution patterns of enzymes in rat-liver tissue. Biochem J 60, 604–617.
3. Novikoff, A.B, Beaufay, H., and de Duve, C. (1956) Electron microscopy of lysosome-rich fractions from rat liver. Journal Biophys Biochem Cytol. 2, 179–190.
4. Clark, S.L. (1957) Cellular differentiation in the kidneys of newborn mice studied with the electron microscope. J Biophys Biochem Cytol 3, 349–376.
5. Novikoff, A.B. (1959) The proximal tubule cell in experimental hydronephrosis. J Biophys Biochem Cytol 6, 136–138.
6. Ashford, T.P., and Porter, K.R. (1962) Cytoplasmic components in hepatic cell lysosomes. J Cell Biol 12, 198–202.
7. Novikoff, A.B., and Essner, E. (1962) Cytolysomes and mitochondrial degeneration. J Cell Biol 15, 140–146.
8. de Duve, C., and Wattiaux, R. (1966) Functions of lysosomes. Annu Rev Physiol 28, 435–492.
9. Behnke, O. (1963) Demonstration of acid phosphatase-containing granules and cytoplasmic bodies in the epithelium of foetal rat duodenum during certain stages of differentiation. J Cell Biol18, 251–265.
10. Bruni, C., and Porter, K.R. (1965) The fine structure of the parenchymal cell of the normal rat liver: I. General observations. Am J Pathol 46, 691–755.
11. Hruban, Z., Spargo, B., Swift, H., Wissler, R.W., and Kleinfeld, R.G. (1963) Focal cytoplasmic degradation. Am J Pathol 42, 657–683.
12. Moe, H., and Behnke, O. (1962) Cytoplasmic bodies containing mitochondria, ribosomes, and rough surfaced endoplasmic membranes in the epithelium of the small intestine of newborn rats. J Cell Biol 13, 168–171.
13. Napolitano, L. (1963) Cytolysomes in metabolically active cells. J Cell Biol 18, 478–481.
14. Bonneville, M.A. (1963) Fine structural changes in the intestinal epithelium of the bullfrog during metamorphosis. J Cell Biol 18, 579–597.
15. Mortimore, G.E., and Schworer, C.M. (1977) Induction of autophagy by amino-acid deprivation in perfused rat liver. Nature 270, 174–176.
16. Pfeifer, U., and Warmuth-Metz, M. (1983) Inhibition by insulin of cellular autophagy in proximal tubular cells of rat kidney. Am J Physiol 244, E109-114.
17. Seglen, P.O., and Gordon, P.B. (1982) 3-Methyladenine: specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc Natl Acad Sci USA 79, 1889–1892.
18. Arstila, A.U., and Trump, B.F. (1968) Studies on cellular autophagocytosis. The formation of autophagic vacuoles in the liver after glucagon administration. Am J Pathol 53, 687–733.
19. Takeshige, K., Baba, M., Tsuboi, S., Noda, T., and Ohsumi, Y. (1992) Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J Cell Biol 119, 301–311.
20. Tsukada, M., and Ohsumi, Y. (1993) Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett 333, 169–174.
21. Klionsky, D.J., Cregg, J.M. Dunn, W.A. Jr., Emr, S.D., Sakia, J., Sandoval, I.V., Sibirnya, Y.A., Subramani, S., Thumm, M., Veenhuis, M., and Ohsumi, Y. (2003) A unified nomenclature for yeast autophagy-related genes. Dev Cell 5, 539-545.
22. Kametaka, S., Matsuura, A., Wada Y., and Ohsumi, Y. (1996) Structural and functional analyses of APG5, a gene involved in autophagy in yeast. Gene 178, 139-43.
23. Funakoshi, T., Matsuura, A., Noda, T., Ohsumi Y. (1997) Analyses of APG13 gene involved in autophagy in yeast, Saccharomyces cerevisiae. Gene. 192, 207-213.
24. Matsuura, A., Tsukada, M., Wada, Y., and Ohsumi, Y. (1997) Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene 192, 245–250.
25. Kamada, Y., Funakoshi, T., Shintani, T., Nagano, K., Ohsumi, M., and Ohsumi, Y. (2000) Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol 150, 1507–1513.
26. Ohsumi, Y. (2014) Historical landmarks of autophagy research. Cell Res 24, 9–23.
27. Kirisako, T., Baba, M., Ishihara, N., Miyazawa, K., Ohsumi, M., Yoshimori, T., Noda, T., and Ohsumi, Y. (1999) Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J Cell Biol 147, 435–446.
28. Mizushima, N., Noda, T., Yoshimori, T., Tanaka, Y., Ishii, T., George, M.D., Klionsky, D.J., Ohsumi, M., and Ohsumi, Y. (1998) A protein conjugation system essential for autophagy. Nature 395, 395–398.
29. Shintani, T., Mizushima, N., Ogawa, Y., Matsuura, A., Noda, T., and Ohsumi, Y. (1999) Apg10p, a novel protein-conjugating enzyme essential for autophagy in yeast. EMBO J 18, 5234–5241.
30. Mizushima, N., Noda, T., and Ohsumi, Y. (1999) Apg16p is required for the function of the Apg12p-Apg5p conjugate in the yeast autophagy pathway. EMBO J 18, 3888–3896.
31. Ichimura, Y., Kirisako, T., Takao, T., Satomi, Y., Shimonishi, Y., Ishihara, N., Mizushima, N., Tanida, I., Kominami, E., Ohsumi, M., et al. (2000) A ubiquitin-like system mediates protein lipidation. Nature 408, 488–492.
32. Hanada, T., Noda, N.N., Satomi, Y., Ichimura, Y., Fujioka, Y., Takao, T., Inagaki, F., and Ohsumi, Y. (2007) The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J Biol Chem 282, 37298–37302.
33. Nakatogawa, H., Ichimura, Y., and Ohsumi, Y. (2007) Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell 130, 165–178.
34. Xie Z., Nair U., Klionsky D.J. (2008) ATG8 controls phagophore expansion during autophagosome formation. Mol Cell Biol 19, 3290-3298.
35. Kabeya, Y., Mizushima, N., Ueno, T., Yamamoto, A., Kirisako, T., Noda, T., Kominami, E., Ohsumi, Y., and Yoshimori, T. (2000) LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J 19, 5720–5728.
36. Mizushima, N., Yamamoto, A., Matsui, M., Yoshimori, T., and Ohsumi, Y. (2004) In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell 15, 1101–1111.
37. Kuma, A., Hatano, M., Matsui, M., Yamamoto, A., Nakaya, H., Yoshimori, T., Ohsumi, Y., Tokuhisa, T., and Mizushima, N. (2004) The role of autophagy during the early neonatal starvation period. Nature 432, 1032–1036.
38. Mizushima, N., and Komatsu, M. (2011) Autophagy: Renovation of cells and tissues. Cell 147, 728-741.
39. Liu, L., Sakakibara, K., Chen, Q., Okamoto, K. (2014) Receptor-mediated mitophagy in yeast and mammalian systems. Cell Res 24, 787-795.
40. Li, W.W., Li, J., Bao, J.K. (2012) Microautophagy: lesser-known self-eating. Cell Mol Life Sci 69, 1125-1136.
41. Cuervo, A.M., and Wong, E. (2014) Chaperone-mediated autophagy: roles in disease and aging. Cell Res 24, 92–104.
42. Liang, X.H., Jackson, S., Seaman, M., Brown, K., Kempkes, B., Hibshoosh, H., and Levine, B. (1999) Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 402, 672–676.
43. Choi, A.M.K., Ryter, S.W., and Levine, B. (2013) Autophagy in human health and disease. N Engl J Med 368, 651–662.
44. Ravikumar, B., Vacher, C., Berger, Z., Davies, J.E., Luo, S., Oroz, L.G., Scaravilli, F., Easton, D.F., Duden, R., O’Kane, C.J., et al. (2004)Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet 36, 585–595.
45. Ravikumar, B., Duden, R., and Rubinsztein, D.C. (2002) Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum Mol Genet 11, 1107–1117.
46. Komatsu, M., Waguri, S., Chiba, T., Murata, S., Iwata, J.-I., Tanida, I., Ueno, T., Koike, M., Uchiyama, Y., Kominami, E., et al. (2006) Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 441, 880–884.
47. Hara, T., Nakamura, K., Matsui, M., Yamamoto, A., Nakahara, Y., Suzuki-Migishima, R., Yokoyama, M., Mishima, K., Saito, I., Okano, H., et al. (2006) Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 441, 885–889.
48. Ebrahimi-Fakhari, D., Saffari, A., Wahlster, L., Lu, J., Byrne, S., Hoffmann, G.F., Jungbluth, H., and Sahin, M. (2016) Congenital disorders
49. Nakagawa, I., Amano, A., Mizushima, N., Yamamoto, A., Yamaguchi, H., Kamimoto, T., Nara, A., Funao, J., Nakata, M., Tsuda, K., et al. (2004) Autophagy defends cells against invading group A Streptococcus. Science 306, 1037–1040.
50. Gutierrez, M.G., Master, S.S., Singh, S.B., Taylor, G.A., Colombo, M.I., and Deretic, V. (2004) Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell 119, 753–766.
Nils-Göran Larsson, MD, PhD
Professor of Mitochondrial Genetics, Karolinska Institutet
Adjunct Member of the Nobel Committee
Member of the Nobel Assembly
Maria G. Masucci, MD, PhD
Professor of Virology, Karolinska Institutet
Adjunct Member of the Nobel Committee
Member of the Nobel Assembly
Illustrations: Mattias Karlén
* Footnotes
The Nobel Prize in Physiology or Medicine 1974 to Albert Claude, Christian de Duve and George E. Palade “for their discoveries concerning the structural and functional organization of the cell”
https://www.nobelprize.org/prizes/medicine/1974/claude/facts/
https://www.nobelprize.org/prizes/medicine/1974/duve/facts/
https://www.nobelprize.org/prizes/medicine/1974/palade/facts/
Glossary of Terms
Lysosome: an organelle in the cytoplasm of eukaryotic cells containing degradative enzymes enclosed in a membrane.
Phagophore: a vesicle that is formed during the initial phases of macroautophagy. The phagophore is extended by the autophagy machinery to engulf cytoplasmic components.
Autophagosome: an organelle that encloses parts of the cytoplasm into a double membrane that fuses to the lysosome where its content is degraded. The autophagosome is the key structure in macroautophagy.
Selective autophagy: a type of macroautophagy that mediates the degradation of specific cytoplasmic components. Different forms of selective autophagy are called mitophagy (degrades mitochondria), ribophagy (degrades ribosomes), lipophagy (degrades lipid droplets) xenophagy (degrades invading microorganisms) etc.
Nobel Prizes and laureates
Six prizes were awarded for achievements that have conferred the greatest benefit to humankind. The 12 laureates' work and discoveries range from proteins' structures and machine learning to fighting for a world free of nuclear weapons.
See them all presented here.
